THE MITOCHONDRIAL GENOME OF TRYPANOSOMES
Research in the Simpson Laboratory
The trypanosomes or kinetoplastid protozoa comprise a large group of
parasitic flagellated cells that are the causal agents for a variety of
human and animal diseases. Research in Dr. Simpson's laboratory has
focused on the molecular biology of the mitochondrial genome in these
organisms from several points of view. These cells represent one of the
earliest eukaryotic lineages containing mitochondria and, as such, possess
many unusual physical and biochemical features, among which is a
mitochondrial genome known as 'kinetoplast DNA' that consists of a network
of thousands of catenated mini- and maxicircles, transcripts of which are
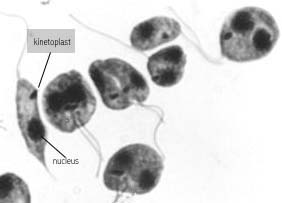
modified by a novel process termed 'RNA editing'. Click here
to see several micrographs of kDNA networks.
The cells can be plated on agar. Click here
to see a movie of T. brucei procyclic colonies on agar.
RNA Editing of Mitochondrial Transcripts in the Mitochondrion of
Trypanosomatids
RNA editing in trypanosomatid protozoa involves the insertion and
deletion of uridine residues (U's) at specific sites within coding regions
of mRNA transcripts of the maxicircle
genome. The sequence information for editing is contained in a class
of small RNAs termed guide RNAs(gRNAs), which were previously discovered
in this laboratory. gRNAs
are small RNAs which, at the 5' end have a region of complementarity with
mRNA sequence just downstream of the sites to be edited, and at the 3' end
have a non-encoded oligo[U] tail. Click here
to see several examples of edited sequence aligned with the cognate
gRNAs.
The majority of the gRNAs are encoded in the thousands of minicircle
molecules which are catenated together into a single giant network of DNA.
This laboratory showed previously that the 3'
to 5' polarity of editing is due to the creation of upstream gRNA
anchor sequences by downstream editing. Here is a figure of the pan-edited
RPS12 gene
Two basic models have been proposed by our laboratory for the mechanism
of editing, one of which involves a cleavage, 3'-terminal U addition, and
religation.
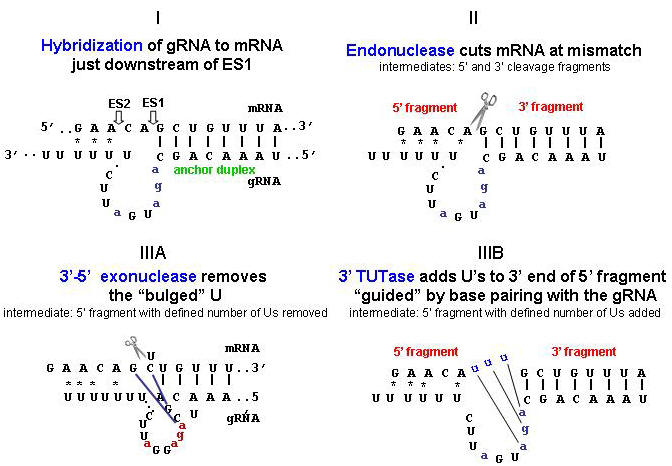
The other,
which was proposed independently by Cech, involves two successive
transesterifications such as occur in RNA splicing, with a transfer of U's
from the 3' end of the gRNA or directly from UTP to the editing site.
gRNA/mRNA chimeric molecules, which were discovered by the Simpson
laboratory, were initially proposed to be intermediates in the
transesterification model, but recent evidence suggests that these
molecules represent non-productive byproducts of the cleavage-ligation
process.
The Evolution of RNA Editing
A rooted phylogenetic tree of the kinetoplastid protozoa was constructed
from nuclear rRNA sequences, which, together with a comparative analysis
of editing of three maxicircle genes in several trypanosomatid species,
led to the surprising conclusion that extensive or pan-editing, mediated
by multiple overlapping guide RNAs, is phylogenetically more ancient than
the limited editing which occurs at the 5' end of editing domains
(5'-editing), and that RNA editing preceded the evolution of parasitism in
this group.
The maxicircle genomic organization
in all analyzed trypanosomatid species is identical, but the extent of
editing varies considerably (Click here
for NAR cover showing a comparison of maxicircle genomes). This can be
visualized in a comparison
of the extent of editing of the A6 or MURF4 mRNA in three species.
The editing appears to become limited to the 5' end of the editing domain.
This can be also seen in a comparison of the editing of the ND7 mRNA in T. brucei
and L.
tarentolae. In T. brucei there is pan-editing of two domains,
whereas in L. tarentolae, editing is limited to the 5' end of each domain.
The phylogenetic analysis also showed that the bodonid/cryptobiids
represent an early diverged sister group to the trypanosomatids, as was
proposed previously by classical analyses. Click
here to see a diagram of the taxonomy of kinetoplastid protozoa.
Analysis of one cryptobiid species, Trypanoplasma borreli, which is
discussed below, showed the presence of the U addition/deletion type of
RNA editing of several mitochondrial transcripts, in spite of a complete
dissimilarity of the mitochondrial gene order.
The evidence indicates that ancestral kinetoplastid cryptogenes were
probably pan-edited and the 5'-edited homologues were possibly generated
by several independent retroposition events from partially edited RNAs. A
comparison of the extent of editing in an old laboratory strain of
Leishmania tarentolae and a recently isolated strain has provided
additional evidence for this mechanism.
This research was done by Dimitri Maslov and Otavio Thiemann.
Disruption of RNA Editing by Prolonged Culture of the
Promastigote Form
RNA editing in kinetoplastids appears to be a labile genetic trait that
is affected by prolonged cell culture. The transcripts of the G1-G5
cryptogenes are pan-edited in the recently isolated LEM125 strain of
Leishmania tarentolae, but not in the UC strain which has been in culture
for 55 years. At least 32 minicircle-encoded guide
RNAs encoding gRNAs for the editing of these transcripts were lost
during the culture history of the old lab strain, probably due to the
absence of a selective pressure for the protein products, which include
subunits of complex I of the respiratory chain. See also guide
RNAs2.
Click here
to see a diagram of the construction of the gRNA library which was used to
detect the additional gRNAs in the LEM125 strain..
The absence of gRNAs for the editing of G5 in the UC strain led to the
existence of extensively misedited RNAs. Some
of these misedited RNAs showed correct editing of block I, and
misediting upstream. The editing of Block I was mediated by a
maxicircle-encoded gRNA, which was present in both strains. Several
non-cognate gRNAs were identified in the UC strain which could account for
specific misedited upstream sequences.
This research was done by Otavio Thiemann.
Diagnosis of Chagas Disease
Trypanosoma cruzi is the causal agent of Chagas Disease, an
important disease of the American tropics, for which there is no adequate
chemotherapy or vaccine. A knowledge of the RNA editing system and the
genetic function of these molecules could aid in development of a rational
chemotherapy for this disease. The previously observed extensive sequence
heterogeneity of the kinetoplast minicircle DNA in Trypanosoma cruzi, both
intra- and inter-strain, has raised the question as to how the minicircle
DNA in this species can have any gRNA-coding capacity at all, since there
does not appear to be any variable region sequences conserved between
different strains. To address this question, the complete edited sequence
of maxicircle unidentified reading frame 4 mRNA was obtained and we were
able to identify 25
cognate gRNAs from gRNA libraries constructed from two clonal
strains of T. cruzi - Sylvio X10/CL1 and CAN III/CL1.
Libraries of PCR-amplified minicircle variable regions were also
constructed for both strains. A single gene for each gRNA was identified
in the same polarity within specific minicircle
variable regions from both strains, 60-100 nucleotides downstream
from the conserved 12mer sequence. GTP-capped
total gRNA from one strain failed to cross-hybridize with minicircle
DNA from the other strain. The explanation for this proved to be the
number of polymorphisms, mainly transitions, within the homologous gRNAs
in the two strains. In most cases these transitions did not destroy the
edited mRNA/gRNA base-pairing, as a result of the allowed G-U wobble
base-pairing. The sequences of the variable regions containing homologous
gRNAs in the two strains probably derived from an ancestral sequence, and
each has accumulated sufficient polymorphisms so as not to allow
hybridization. Within a strain, multiple redundant gRNAs were identified
which encode identical editing information but have different sequences.
Dr. Simpson's laboratory previously developed a sensitive diagnostic
test for the presence of these parasites in chronically ill
patients by PCR amplification of parasite-specific but highly polymorphic
DNA fragments from kinetoplast minicircle molecules. A single pair of PCR
primers within a conserved region of the mini-exon repeat was used to
amplify the repeats from 10 species of pathogenic Leishmania belonging to
4 major clinical groups and also from 3 species of Trypanosoma.
Oligonucleotide hybridization probes for the detection and
identification of the PCR-amplified mini-exon repeats were constructed
from alignments of mini-exon intron and intergenic sequences. The probes
generated from mini-exon intergenic regions of the L. (V.) braziliensis,
L. (L.) donovani and L. (L.) mexicana species hybridized specifically to
their cognate groups without discriminating between the species within the
groups. The probes for L. (L.) major and L. (L.) aethiopica were
species-specific, while the L. (L.) tropica probe also hybridized with the
L. (L.) aethiopica mini-exon repeat. The mini-exon intron-derived probes
for T. cruzi, T. rangeli and T. brucei were species-specific. This method
involving the detection of specific PCR-amplified products produced using
a single primer set represents a novel sensitive and specific assay for
multiple trypanosomatid species and groups. It essentially represents a
multiplex assay and should complement the minicircle-based diagnostic
assay previously developed by this laboratory.
Here is
an article on the history of the development of this assay.
This research was done by Herbert Avila et al.
RNA Editing in Crithidia fasciculata
Although the mitochondrial uridine insertion/deletion, guide
RNA-mediated type of RNA editing has been described in Crithidia
fasciculata, no evidence for the encoding of guide RNAs in the kinetoplast
minicircle DNA has been presented. There has also been a question as to
the capacity of the minicircle DNA in this species to encode the required
variety of gRNAs, since the kinetoplast DNA from the C1 strain has been
reported as essentially containing a single minicircle sequence class. To
address this problem, the genomic and mature edited sequences of the MURF4
and RPS12
cryptogenes were determined, and a gRNA library was constructed from
mitochondrial RNA. Five specific gRNAs were identified, two of which edit
blocks within the MURF4 mRNA, and three of which edit blocks within the
RPS12 mRNA. The genes
for these gRNAs are all localized with identical polarity within one of
the two variable regions of specific minicircle molecules, approximately
60 bp from the 'bend' region. These minicircles were found to represent
minor sequence classes representing approximately 2% of the minicircle DNA
population in the network. The major minicircle sequence class also
encodes a gRNA at the same relative genomic location, but the editing role
of this gRNA was not determined. These results confirm that kinetoplast
minicircle DNA molecules in this species encode gRNAs, as is the case in
other trypanosomatids, and suggest that the copy number of specific
minicircle sequence classes can vary dramatically without an overall
effect on the RNA editing system.
This research was done by Shinji Yasuhira.
Guide RNAs and guide RNA genes in the cryptobiid
kinetoplastid protozoan, Trypanoplasma borreli
Trypanoplasma borreli belongs to the bodonid/cryptobiid group of
kinetoplastid protozoa, which represents a sister group to the
trypanosomatids. RNA transcripts from several mitochondrial genes in this
organism undergo the trypanosomatid type of uridine addition/deletion RNA
editing. A gRNA cDNA library was constructed and
five gRNAs were identified, one for editing the ribosomal protein
S12 mRNA, three for editing the cytochrome oxidase subunit I mRNA and one
for editing the cytochrome b mRNA. All of the gRNAs contained non-encoded
oligo[U] sequences at their 5' end, as well as at the 3' end as is common
with gRNAs in trypanosomatids. The mechanism for addition of the 5'
non-encoded oligo[U] sequence and the function of this sequence are
unknown. The T. borreli gRNAs were shorter (25-35 nt, excluding the 5'
oligo[U]) than gRNAs in trypanosomatids (45-50 nucleotides), indicating a
smaller size of editing blocks in this organism. Genomic sequences for two
gRNAs were cloned and sequenced. These two gRNA-encoding sequences were
shown to originate from the 170-200 kb Component I molecules which
represent a possible homologue of minicircle DNA in trypanosomatids, and
not from the 80 kb Component II molecules which contain the structural
genes and cryptogenes.
This research was done by Shinji Yasuhira
Phylogenetic affinity of mitochondria of Euglena gracilis
and kinetoplastids using cytochrome oxidase I and hsp60.
The mitochondrial DNA-encoded cytochrome oxidase subunit I (COI) gene
and the nuclear DNA-encoded hsp60 gene from the euglenoid protozoan
Euglena gracilis were cloned and sequenced. The COI sequence represents
the first example of a mitochondrial genome-encoded gene from this
organism. This gene contains seven TGG tryptophan codons and no TGA
tryptophan codons, suggesting the use of the universal genetic code. This
differs from the situation in the mitochondrion of the related
kinetoplastid protozoa, in which TGA codes for tryptophan. In addition, a
complete absence of CGN triplets may imply the lack of the corresponding
tRNA species. COI cDNAs from E. gracilis possess short 5' and 3'
untranslated transcribed sequences and lack a 3' poly[A] tail. The COI
gene does not require uridine insertion/ deletion RNA editing, as occurs
in kinetoplastid mitochondria, to be functional, and no short guide
RNA-like molecules could be visualized by labeling total mitochondrial RNA
with [alpha-32P]GTP and guanylyl transferase. In spite of the differences
in codon usage and the 3' end structures of mRNAs, phylogenetic analysis
using the COI
and hsp60
protein sequences suggests a monophyletic relationship between the
mitochondrial genomes of E. gracilis and of the kinetoplastids, which is
consistent with the phylogenetic relationship of these groups previously
obtained using nuclear ribosomal RNA sequences.
This research was done by Shinji Yasuhira.
Involvement of mitochondrial ribonucleoprotein complexes
in RNA Editing
By analysis of mitochondrial extracts on glycerol gradients, two types
of ribonucleoprotein (RNP) complexes were identified containing gRNAs. The
T-class sediments at approximately 10S and consists of approximately six
complexes, the endogenous RNA of which can be self-labeled with [32P]UTP.
The most abundant T-complex, T-IV, was visualized by electron microscopy
as 980-140 A particles. This complex exhibits terminal uridylyl
transferase (TUTase) activity and contains gRNAs. The other T-complexes
also contain mRNA fragments. The function of T-IV may be to add U's to the
3' end of the gRNA. The second class of RNP complexes consists of 170-300
A particles which show little TUTase activity, and exhibit an in vitro RNA
editing-like activity.
Two mitochondrial proteins of 18 kD and 51 kD which appeared to show
interaction with exogenous gRNAs were isolated and the genes cloned. These
proteins were components of complex T-I and T-VI, respectively. The
proteins possess 17 and 9 amino acid N terminal cleavable mitochondrial
targeting sequences. The p18 protein localized throughout the entire
tubular mitochondrion. and has been identified by the Benne lab as a
subunit of the mtochondrial ATPase. The p51 protein was identified as a
mitochondrial aldehyde dehydrogenase. In addition, 71 kD and 62 kD
proteins which comigrated in native gels with other T-complexes were
identified as hsp70 and hsp60 homologs.
The majority of the protein-RNA interactions in the T-complexes were
shown to be low affinity. However, digestion of the extract with
micrococcal nuclease and saturation with rRNA was found to uncover a high
affinity RNP complex, which involves at least two proteins interacting
with the endogenous gRNAs.
The second class of complexes sediments at 20S. A gRNA-independent in
vitro editing-like activity which comigrates with the 20S complexes was
assayed by following the incorporation of radioactively labeled U's into
the pre-edited region of a synthetic mRNA substrate by digestion with
RNase H and specific oligonucleotides. The incorporation is limited
precisely to the pre-edited region and is dependent on some type of
endogenous RNA, as shown by micrococcal nuclease digestion experiments.
Labeled C residues were incorporated into the same sites as U residues,
but at a 20 fold lower level.
This research was done by Marian Peris, Agda Simpson, Frederic Bringaud,
Elaine Byrne and Georges Frech.
Native gel analysis of ribonucleoprotein complexes from a
Leishmania tarentolae mitochondrial extract
Two polypeptides of 50 and 45 kDa were adenylated by incubation of a
mitochondrial extract from Leishmania tarentolae with [a-32P]ATP. These
proteins were components of the 20S complex, which migrated as a single
band of approximately 1800 kDa in a native gel. The facts that RNA ligase
activity cosedimented at 20S and that the ATP-labeled p45 and p50
polypeptides were deadenylated upon incubation with a ligatable RNA
substrate suggests that these proteins may represent charged intermediates
of a mitochondrial RNA ligase. Hybridization of native gel blots with
guide RNA (gRNA) probes showed the presence of gRNA in the previously
identified T-IV complexes that sedimented in glycerol at 10S and contained
TUTase activity, and also in a previously unidentified class of
heterodisperse complexes that sedimented throughout the gradient. gRNAs
were not detected in the p45+p50-containing complex. The heterodisperse
gRNA-containing complexes were sensitive to incubation at 27oC and appear
to represent complexes of T-IV subunits with mRNA. Polyclonal antiserum to
a 70 kDa protein that purified with terminal uridylyl transferase activity
was generated, and the antiserum was used to show that this p70
polypeptide was a component of both the T-IV and the heterodisperse
gRNA-containing complexes. We propose that the p45+p50-containing 1800 kDa
complex and the p70+gRNA -containing heterodisperse complexes interact in
the editing process.
This research was performed by Marian Peris, Agda M. Simpson, Jeremy
Grunstein, Joanna E. Liliental and Georges C. Frech
Guide RNA-independent RNA editing in vitro
A mitochondrial extract from Leishmania tarentolae directs the
incorporation of uridylate (U) residues within the pre-edited domain of
synthetic cytochrome b (CYb) and NADH dehydrogenase subunit 7 mRNA. This
has several characteristics of an in vitro RNA editing activity, but no
direct evidence for involvement of guide RNAs was obtained. In fact,
evidence obtained by Greg Connell indicates that this activity is
independent of both exogenous and endogenous gRNAs. The incorporation is
limited to the preedited region but the number and localization of U's
inserted is not identical to that found in mature correctly edited RNA.
The activity is selectively inhibited by digestion of the lysate with
micrococcal nuclease, possibly suggesting a requirement for some type of
endogenous RNA. A low level of incorporation of [alpha-32P]CTP occurs at
the same sites as UTP.
Stereochemical evidence for the enzyme cascade model for
RNA editing
Chiral phosphorothioates were used to investigate the
stereoconfiguration requirements and the stereochemical course of an RNA
editing-like internal uridine (U)-incorporation activity and a 3' terminal
U-addition activity using a mitochondrial extract from L. tarentolae. The
extract utilizes SP-[a-S]UTP for both 3' and internal U- incorporation
into substrate RNA. The internal as well as the 3' incorporation of
SP-[a-S]UTP proceeds via inversion
of the stereoconfiguration. The mitochondrial RNA ligase
produces an inversion of the stereoconfiguration. Iternal U-incorporation
does not occur at sites containing thiophosphodiesters of the RP
configuration. The results
are compatible with an enzyme cascade model for this in vitro U-insertion
activity involving sequential endonuclease, uridylyl transferase directly
from UTP and RNA ligase steps, and incompatible with models involving the
transfer of U residues from the 3' end of gRNAs.
This research was done by Georges Frech.
The role of mRNA structure in guide RNA-independent RNA
editing in vitro
A primer extension assay
was used for the detection of uridine insertions occurring in vitro in
synthetic pre-edited cytochrome b mRNA during incubation with a Leishmania
tarentolae mitochondrial extract. Two different activities were detected
that inserted uridines within the first two editing sites: one
that is dependent on the secondary structure of the mRNA but is
independent of both exogenous and endogenous guide RNA, and a second that
does not put the same structural constraints on the mRNA, but is dependent
on the presence of a cognate guide RNA. The possibility that contaminating
gRNA-mRNA chimeric molecules were the source of the extension ladders was
eliminated by performing the extension assay on RNAs transcribed from
3'-tagged RT-PCR-amplified cDNAs. Leaving aside the question of the
biological relevance of the gRNA-independent reaction, it is possible that
the structure of the RNAs supporting this in vitro reaction could be
mimicking the RNA structures occurring during gRNA-mediated editing. For
example, the duplex
formed by foldback of the Cyb mRNA 5' and 3' extensions, which is required
for the gRNA-independent reaction, may serve the same function as the
anchor duplex created by the gRNA binding to the mRNA. Annealing of the
cognate gRNA containing a complementary anchor sequence with the
5'-extended mRNA construct would be predicted to disrupt the mRNA
secondary structure, thereby accounting for the observed inhibitory effect
of the addition of exogenous gRNA on the gRNA-independent U-insertion
activity. The creation of a recognition element for the assembly of the
U-insertion machinery may represent an important role of the gRNA in the
gRNA-mediated reaction. The recognition element could be the double
stranded RNA formed by the gRNA-mRNA interaction that may be mimicked by
the intramolecular helix necessary for the gRNA-independent reaction.
This work was performed by Greg Connell and Elaine Byrne.
Guide RNA-directed uridine-insertion RNA editing in vitro
Guide RNAs (gRNAs) have been proposed to mediate uridine (U)
addition/deletion editing of mitochondrial messenger RNAs in kinetoplastid
protozoa. The U's are proposed to be derived either from UTP by two
successive cleavage-ligations or transesterifications, or from the 3' end
of the gRNA by the same mechanisms. By use of a sensitive and specific
primer extension assay,
we have demonstrated guide RNA-dependent U-insertions
into a specific editing site of a preedited mRNA which was incubated in a
mitochondrial extract from Leishmania tarentolae. The predominant number
of U-insertions was determined by the number of guiding nucleotides in the
added gRNA, and the formation of a gRNA-mRNA anchor duplex was necessary
for activity. UTP and a-b bond hydrolysis of ATP were required, and the
activity was inhibited above 50-100 mM KCl. A guide RNA-independent
insertion of up to approximately 13 U's occurred in the absence of the
added cognate gRNA; the extent of this activity was affected by sequences
upstream and downstream of the edited region. Heparin inhibited the guide
RNA-independent U-insertion activity and had no effect on the guide
RNA-dependent activity. Blocking the 3'OH of the gRNA had little effect on
the gRNA-dependent U-insertion activity. The data are consistent with a
modified enzyme cascade model
in which the U's are derived directly from UTP.
This work was performed by Elaine Byrne and Greg Connell.
Sequence-dependent importation of tRNAs into the
mitochondrion of Leishmania tarentolae
Another topic of research in the Simpson lab is the importation of tRNAs
into the kinetoplast- mitochondrion of L. tarentolae. Previous research
had shown that no mitochondrial tRNAs were encoded in the maxicircle (or
minicircle) genome. Therefore the mitochondrial tRNAs had to be derived
from the cytosol and be encoded in nuclear genes. To investigate this, an
in vivo transfection method was employed. tRNAIle(UAU) was shown
previously by Shi et al., 1994 and Lye et al., 1993 to exclusively
localized within the mitochondrion and tRNAGln(CUG) exclusively in the
cytosol. L. tarentolae cells were transfected with plasmids encoding
either tRNAIle or tRNAGln that were tagged with altered sequences in the D
loop, permitting discrimination from the endogenous tRNAs. Primer
extension analysis was used to show that the plasmid-encoded genes were
expressed and that the tagged tRNAs showed a similar intracellular
localization as the endogenous tRNAs. Exchange or deletion of the
5'-flanking genomic sequences had no effect on the expression or
mitochondrial localization of the tagged tRNAIle or on the expression or
cytosolic localization of the tagged tRNAGln, suggesting that the signals
for importation are localized within the tRNA itself. Swapping the D
loop+stem from the exclusively cytosolic tRNAGln with that from the
tRNAIle produced a partial mitochondrial localization of the
plasmid-expressed mutated tRNAGln. However, D loop exchange did not
eliminate the mitochondrial localization of the plasmid-expressed mutated
tRNAIle, suggesting that tertiary structure or additional sequence
elements may be involved in the importation signal.
This research was done by Beatriz Lima.
The mitochondrial glutamate dehydrogenase from Leishmania
tarentolae is a guide RNA-binding protein
To identify specific proteins interacting with guide RNAs in
mitochondrial ribonucleoprotein complexes from Leishmania tarentolae,
fractionated and unfractionated mitochondrial extracts were subjected to UV cross-linking
with added labeled gRNA and also with [a-32P]UTP-labeled
endogenous RNA. An abundant 110-kD protein (p110) localized in the T-V
complex, which sediments in glycerol gradients
at the leading edge of the 10S terminal uridylyl transferase peak, was
found to interact with both types of labeled RNAs. The p110 protein was
gel-isolated and subjected to microsequence analysis, and the gene was
cloned. The sequence
revealed significant similarity with mitochondrial glutamate
dehydrogenases. A polyclonal antiserum was raised against a recombinant
fragment of the p110 gene and was used to demonstrate a stable and
specific gRNA-binding activity by co-immunoprecipitation
and competitive gel-shift analyses.
Complex formation was strongly inhibited by competition with poly[U] or by
deletion or substitution
of the gRNA 3'-oligo[U] tail. Also, addition
of a 3' oligo[U] tail to an unrelated transcript was sufficient for p110
binding. Both the gRNA-binding activity of the p110 protein and in vitro
gRNA-independent and gRNA-dependent uridine insertion activities in the
mitochondrial extract were inhibited by high concentrations of dinucleotides.
This work was performed by Frédéric Bringaud, Renata Stripecke, Georges
C. Frech, Stephen Freedland, Christoph W. Turck and Elaine M. Byrne.
Knockout of the glutamate dehydrogenase gene in
bloodstream Trypanosoma brucei in culture has no effect on editing
of mitochondrial mRNAs
Glutamate dehydrogenase (GDH) was shown previously to bind the 3'
oligo[U] tail of the mitochondrial guide RNAs (gRNAs) of Leishmania
tarentolae, apparently in the dinucleotide pocket. Bloodstream Trypanosoma
brucei cells in culture represent a good system to investigate the
genetic effects of knocking out kinetoplastid nuclear genes to test a role
in RNA editing, since editing of several mitochondrial genes occurs but is
dispensable for viability. Both GDH alleles of bloodstream T. brucei in
culture were replaced by drug resistant markers without any effect on
viability. The ratios of edited to unedited mRNAs for several cryptogenes
were assayed by primer extension analysis. The steady state abundances of
these edited RNAs were unaffected by the double knockout. This evidence
suggests that GDH may not play a role in the editing reaction in
bloodstream trypanosomes in culture, but this conclusion is tentative
since there could be redundant genes for any biological function. We
employed a double allelic replacement technique to generate a tetracycline
inducible conditional expression of an ectopic copy of the deleted gene in
bloodstream trypanosomes in culture. We used this strategy for genes
encoding mitochondrial proteins which are not required during this stage
of the life cycle, but as a general strategy it should be appropriate for
generation of conditional null mutants for essential genes as well.
This work was performed by A. Estevez, F. Kierzenbaum, E. Wirtz, F.
Bringaud and J. Grunstein.
The mitochondrion in dividing Leishmania tarentolae cells
is symmetric and circular and becomes a single asymmetric tubule in
non-dividing cells due to division of the kinetoplast portion
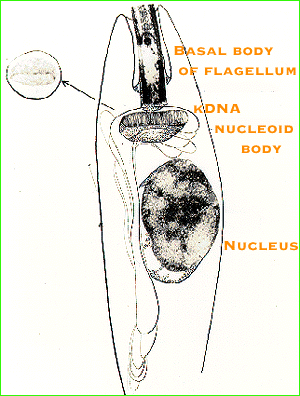
Kinetoplastid protozoa have a single
mitochondrion that extends throughout the cell. The disk-shaped
portion of the mitochondrion adjacent to the basal body of the flagellum
contains the kinetoplast DNA nucleoid body which consists of thousands of
catenated minicircles and a smaller number of catenated maxicircles. The
maxicircles contain structural genes and cryptogenes, rRNA genes, and a
few guide RNA genes The minicircles contain the majority of the guide RNA
genes. The long slender non-dividing stationary phase Leishmania
tarentolae cells in culture have an asymmetric
mitochondrion that consists of a single tubule extending from one
edge of the kinetoplast portion. This presents a problem for cell
division, in that one daughter cell will receive significantly less
mitochondrial membranes than the other cell. We show in this paper that
the solution to this problem is that dividing cells, which are normally
shorter and rounder than stationary phase cells, possess a symmetric circular
mitochondrion that has mitochondrial tubules extending from both
edges of the kinetoplast which are joined in the posterior region of the
cell. This implies that growth of the mitochondrion occurs after cell
division, either from elongation of the longitudinal tubule towards the
anterior of the cell, or from elongation of the kinetoplast portion of the
mitochondrion towards the posterior region and fusion of the tubules.
This work was performed by Larry Simpson and Frank Kretzer.
Lack of evidence for presence of 5' extentions on
tRNAs imported into the mitochondrion of Leishmania
All mitochondrial tRNAs in kinetoplastid protozoa are encoded in nuclear
DNA and transported into the mitochondrion (Simpson et al., 1989, Nucl.
Acids Res. 17: 5427-5445). It has been proposed by Hancock et al. (J.
Biol. Chem., 1990, 265: 19208-19215) that tRNAs in these cells are
imported into the mitochondrion as 5'-extended precursors which are
processed by a mitochondrial RNase P-like activity. We have examined this
hypothesis by cloning and sequencing primer extension products of
mitochondrial tRNAs from L. tarentolae and T. brucei, and have found that
these are derived from circularized mature tRNA molecules. We suggest that
these molecules are produced by the endogenous RNA ligase activity either
in vivo or during mitochondrial isolation. We did not obtain any evidence
for the existence of high molecular weight precursors of mitochondrial
tRNAs. This negative result is consistent with previous in vivo
transfection studies with both L. tarentolae (Lima and Simpson, 1996, RNA
2: 429-440) and T. brucei (Hauser and Schneider, 1995, EMBO J. 14:
4212-4220), in which mitochondrial targeting of plasmid-expressed tRNAs
was independent of the presence of 5'-flanking sequences. We conclude that
the hypothesis for 5'-extended tRNA precursors in kinetoplastid
mitochondrial importation remains to be verified.
This work was performed by Ruslan Aphasizhev.
Cloning and characterization of the Leishmania
tarentolae adenine phosphoribosyltransferase
Adenine phosphoribosyltransferase (APRT) is an important enzyme involved
in the recycling of purine nucleotides in all cells. Parasitic protozoa of
the order Kinetoplastida are unable to synthesize purines de novo and
utilize the salvage pathway for synthesis of ribonucleotides. The aprt
gene was cloned from a Leishmania tarentolae genomic library and
the sequence determined. The L. tarentolae aprt gene contains a
708 nucleotide open reading frame that encodes a 25 kDa protein. The
predicted amino acid sequence
has 85% identity to the APRT of L. donovani (Allen,T., Hwang, H.,
Wilson, K., Hanson, S., Jardim, A. and Ullman, B. (1995) Mol. Biochem.
Parasitol. 74, 99-103). A recombinant protein was expressed in Escherichia
coli, purified
to homogeneity and found to retain enzymatic activity. The L.
tarentolae APRT is active as a homodimer in solution. The
availability of the APRT enzyme from another kinetoplastid protozoan and
the possibility of expressing the recombinant protein in large quantities
should provide the basis for a functional and structural analysis of this
enzyme which has been suggested as a potential target for rational drug
design.
This work was performed by Otavio Thiemann.
Purification and Characterization of MAR1: a
Mitochondrial Associated Ribonuclease from Leishmania tarentolae
A relatively thermostable 22 kDa endoribonuclease (MAR1) was purified
more than 10,000 fold from a mitochondrial extract of Leishmania
tarentolae and the gene cloned. The purified nuclease has a Km of
100-145 ± 33 nM and a Vmax of 1.8-2.9 ± 2 nmoles/min, depending on the RNA
substrate, and yields a 3' OH and a 5' phosphate. Cleavage was limited to
several specific sites in the substrate RNAs tested, but cleavage of
pre-edited RNAs was generally independent of the addition of cognate guide
RNA. The MAR1 gene was expressed in Escherichia coli or in Leishmania
tarentolae cells and the recombinant protein was affinity-purified.
The cleavage specificity of the recombinant enzyme from Leishmania
tarentolae was identical to that of the native enzyme. The single
copy MAR1 gene maps to an 820 kb chromosome and contains an open reading
frame of 579 nt. The 18 amino acid N-terminal sequence shows
characteristics of an uncleaved mitochondrial targeting sequence. Database
searching revealed two homologues of MAR1 corresponding to unidentified
open reading frames in Caenorhabditis elegans (Z69637) and Archaeoglobus
fulgidus (AE000943). The function of MAR1 in mitochondrial RNA
metabolism in L. tarentolae remains to be determined. Click here
and here
to see the figures from this paper.
This work was performed by Juan Alfonzo and Otavio Thiemann.
Phylogenetic Affinities of Diplonema within the
Euglenozoa as Inferred from the SSU rRNA Gene and Partial COI Protein
Sequences
In order to shed light on the phylogenetic position of diplonemids
within the phylum Euglenozoa, we have sequenced small subunit rRNA (SSU
rRNA) genes from Diplonema (syn. Isonema) papillatum and
Diplonema sp. We have also analyzed a partial sequence of the
mitochondrial gene for cytochrome c oxidase subunit I from D.
papillatum. With both markers, the maximum likelihood method favored
a closer grouping of diplonemids with kinetoplastids, while the parsimony
and distance suggested a closer relationship of diplonemids with
euglenoids. In each case, the differences between the best tree and the
alternative trees were small. The frequency of codon usage in the partial
D. papillatum COI was different from both related groups; however,
as is the case in kinetoplastids but not in Euglena, both the
non-canonical UGA codon and the canonical UGG codon were used to encode
tryptophan in Diplonema.
This research was performed by Dmitri Maslov and Shingi Yasuhira.
In vitro uridine insertion RNA editing mediated by
cis-acting guide RN
Guide RNAs involved in mediating RNA editing in vivo are provided in
trans except for the CO2 mRNA, which has the gRNA in cis at
the 3' end of the RNA. We have found that a cognate gRNA provided in
cis at the 3' end of a pre-edited NADH dehydrogenase 7 (ND7) mRNA
substrate can direct U insertions at editing site 1 when incubated with a
mitochondrial lysate from Leishmania tarentolae. The efficiency of
gRNA-dependent U insertion mediated by a cis-acting is greater on a molar
basis than that for a trans-acting gRNA, as expected for a unimolecular
gRNA:mRNA interaction. Blocking the 3' end of a cis-acting gRNA lacking a
3' oligo[U] tail has no effect on gRNA-dependent U insertions, nor does
providing the gRNA in cis upstream of the mRNA, confirming the
previous observation that the terminal 2'- and 3'-hydroxyls of the gRNA
are not involved in U insertion activity. These results also establish
that the oligo[U] tail is not required for U insertion in vitro.
Increasing the extent of base pairing between the 3' end of the gRNA and
the 5' end of the mRNA significantly increases in vitro
gRNA-dependent U insertion at site 1, presumably by maintaining the mRNA
5' cleavage fragment within the editing complex. We speculate that, in
vivo, protein:RNA and/or protein:protein interactions may be
responsible for maintaining the mRNA 5' cleavage fragment in close
proximity to the mRNA 3' cleavage fragment, and that such interactions may
be rate limiting in vitro.
This work was done by Steve Kapuschoc.
The Mitochondrial RNA ligase from Leishmania
tarentolae can join RNA molecules bridged by a complementary RNA
A biochemical characterization was performed with a partially purified
RNA ligase from isolated mitochondria of Leishmania tarentolae.
This ligase has a Km of 25 ± 0.75 nM and a Vmax of 1.0 x10-4 ± 2.4 x 10-4
nmoles/min when ligating a nicked double stranded RNA substrate. Ligation
was negatively affected by a gap between the donor and acceptor
nucleotides. The catalytic efficiency of the circularization of a single
stranded substrate was five-fold less than that of the ligation of a
nicked substrate. These properties of the mitochondrial RNA ligase are
consistent with an expected in vivo role in the process of uridine
insertion/deletion RNA editing, in which the mRNA cleavage fragments are
bridged by a cognate guide RNA.
Click here to
see the figures from this paper.
This work was done by Valerie Blanc, Juan D. Alfonzo, and Ruslan
Aphasizhev.
T7 RNA polymerase-driven transcription in mitochondria of
Leishmania tarentolae and Trypanosoma brucei.
The study of RNA editing and other molecular processes in the
trypanosome mitochondrion would benefit greatly from the ability to insert
and express exogenous DNA in the organelle. However, even with a method to
introduce DNA, the current lack of knowledge about mitochondrial
transcription would hinder efforts to obtain expression. To circumvent
this problem, we have transfected Leishmania tarentolae promastigotes and
Trypanosoma brucei procyclic cells with bacteriophage T7 RNA polymerase
targeted to the mitochondrion. Mitochondria isolated from the
transfectants contained active T7 RNA polymerase, as shown by a
comigration in density gradients of mitochondrial marker enzymes and T7
polymerase activity. A DNA cassette under T7 control was introduced into
isolated mitochondria from the transfectants by electroporation and the
DNA was shown to be transcribed. This system should allow the
transcription of foreign genes of choice within the mitochondrial matrix
either in a transient assay using electroporation of DNA into isolated
mitochondria, or in a stable assay using cells transfected with DNA by the
biolistic gun method.
Click
here to see the figures from this work.
This work was done by Antonio M. Estévez, Otavio H. Thiemann and Juan D.
Alfonzo.
C to U editing of Anticodon of Imported Mitochondrial
tRNATrp allows Decoding of UGA Stop Codon in Leishmania  tarentolae
tarentolae
All mitochondrial tRNAs in kinetoplastid protists are encoded in the
nucleus and imported into the organelle. The tRNATrp(CCA) can decode the
standard UGG tryptophan codon but cannot decode the mitochondrial UGA
tryptophan codon. We show that the mitochondrial tRNATrp undergoes a
specific C to U nucleotide modification in the first position of the
anticodon which allows decoding of mitochondrial UGA codons as tryptophan.
Functional evidence for the absence of a UGA suppressor tRNA in the
cytosol, using a reporter gene, was also obtained, which is consistent
with a mitochondrial localization of this editing event. Leishmania
cells have dealt with the problem of a lack of expression within the
organelle of this non-universal tRNA by compartmentalizing an editing
activity which modifies the anticodon of the imported tRNA.
This work was performed by Juan D. Alfonzo, Valerie Blanc, Antonio M.
Estévez and Mary Anne T. Rubio.
Evolution of RNA Editing in Trypanosome Mitochondria
Two different RNA editing systems have been described in the
kinetoplast-mitochondrion of trypanosomatid protists. The first involves
the precise insertion and deletion of U residues mostly within the coding
regions of maxicircle-encoded mRNAs to produce open reading frames. This
editing is mediated by short overlapping complementary guide RNAs (gRNAs)
encoded in both the maxicircle and the minicircle molecules, and involves
a series of enzymatic cleavage-ligation steps. The second editing system
is a C34 to U34 modification in the anticodon of the imported tRNATrp,
thereby permitting the decoding of the UGA stop codon as tryptophan.
U-insertion editing probably originated in an ancestor of the
kinetoplastid lineage and has evolved in some cases by the replacement of
the original pan-edited cryptogene with a partially edited cDNA. The
driving force for this retroposition was postulated to be the stochastic
loss of entire minicircle sequence classes and their encoded gRNAs upon
segregation of the single kinetoplast DNA network into daughter cells at
cell division. A large plasticity in frequencies of minicircle sequence
classes has been observed during cell culture in the laboratory. Computer
simulations provide theoretical evidence for this plasticity if a random
distribution and segregation model of minicircles is assumed. The specific
C to U tRNA editing probably evolved after the loss of the original
endogenous tRNA genes from the mitochondrial genome and the development of
a mechanism for importation of nuclear-encoded tRNAs into the organelle.
The relationship of the two editing systems is discussed.
This work was performed by Otavio Thiemann, Nickloas Savill, Juan
Alfonzo, and Dmitri Maslov. It was presented at the Colloquium on
Variation and Evolution in Plants and Microorganisms, Jan. 27-29, 2000,
Beckman Center of the National Academies of Sciences and Engineering,
Irvine, CA.
Selective Importation of RNA into Isolated Mitochondria
from Leishmania tarentolae
All mitochondrial tRNAs in kinetoplastid protozoa are encoded in the
nucleus and imported from the cytosol. Incubation of two in vitro
transcribed tRNAs, tRNAIle(UAU) and tRNAGln(CUG),
with isolated mitochondria from Leishmania tarentolae, in the
absence of any added cytosolic fraction, resulted in a protease-sensitive,
ATP-dependent importation, as measured by nuclease protection. Evidence
that nuclease protection represents importation was obtained by the
finding that Bacillus subtilis pre-tRNAAsp was
protected from nuclease digestion and was also cleaved by an
intra-mitochondrial RNase P-like activity to produce the mature tRNA. The
presence of a membrane potential is not required for in vitro importation.
A variety of small synthetic RNAs were also found to be efficiently
imported in vitro. The data suggest that there is a structural requirement
for importation of RNAs greater than approximately 17 nt, and that smaller
RNAs are apparently non-specifically imported. The signals for importation
of folded RNAs have not been determined, but the specificity of the
process was illustrated by the higher saturation level of importation of
the mainly mitochondria-localized tRNAIle as compared to the
level of importation of the mainly cytosol-localized tRNAGln.
Furthermore, exchanging the D-arm between the tRNAIle and the
tRNAGln resulted in a reversal of the in vitro importation
behavior and this could also be interpreted in terms of tertiary structure
specificity.
This work was performed by Mary Anne T. Rubio, Xuan Liu, Harumi Yuzawa
and Juan D. Alfonzo.
End processing precedes mitochondrial importation and
editing of tRNAs in Leishmania tarentolae
All mitochondrial tRNAs in Leishmania tarentolae are encoded in the
nuclear genome and imported into the mitochondrion from the cytosol. One
imported tRNA (tRNATrp) is edited by a C to U modification at the first
position of the anticodon. In order to determine the in vivo substrates
for mitochondrial tRNA importation as well as tRNA editing, we examined
the subcellular localization and extent of 5'- and 3'-end maturation of
tRNATrp(CCA), tRNAIle(UAU), tRNAGln(CUG), tRNALys(UUU), and tRNAVal(CAC).
Nuclear, cytosolic and mitochondrial fractions were obtained with little
cross contamination, as determined by northern analysis of specific marker
RNAs. The tRNAGln was mainly cytosolic in localization, the tRNAIle and
tRNALys were mainly mitochondrial, and the tRNATrp and tRNAVal were shared
between the two compartments. 5'- and 3'-extended precursors of all five
tRNAs were present only in the nuclear fraction, suggesting that the
mature tRNAs represent the in vivo substrates for importation into the
mitochondrion. Consistent with this model, T7-transcribed mature tRNAIle
undergoes importation in vitro into isolated mitochondria more efficiently
than the 5'-extended precursor tRNAIle. The 5'-extended precursor tRNATrp
was found to be unedited, which is consistent with a mitochondrial
localization of this editing reaction. T7-transcribed unedited tRNATrp was
imported in vitro more efficiently than edited tRNATrp, suggesting the
presence of importation determinants in the anticodon.
This research was performed by Steve Kapushoc, Juan Alfonzo and Mary
Anne Rubio.
Uridine insertion/deletion RNA editing in L.
tarentolae mitochondria shows cell cycle dependence
L. tarentolae cells were synchronized using hydroxyurea and the
relative abundance of edited and pre-edited transcripts for 4 maxicircle
genes was analyzed by primer extension. The primers in each case
hybridized to unedited sequence downstream of the editing domain, so that
both unedited and edited extension products can be observed on the same
gel. The FE/UE ratio was found to vary from 1.3 to 2.0 fold for all four
genes during the cell cycle. The ratio peaked in S+G1 and then again in
the same phase of the second synchronized cycle. This variation is most
likely due to variation in the extent of editing. The level of this
regulation is not known. This phenomenon may have some relationship to the
synchronicity of nuclear and mitochondrial replication phases, which
appears to involve a differential expression of nuclear-encoded
replication proteins due to differential turnover of mRNAs (see papers
from Dan Ray lab).
This work was performed by R. Carrillo, O. Thiemann and J. Alfonzo.
Isolation and Characterization of a U-specific 3'-5'
Exonuclease from Mitochondria of Leishmania tarentolae
We have purified a 3'-5' exoribonuclease from mitochondrial extract of
Leishmania tarentolae over 4000-fold through six column fractionations.
This enzyme digested RNA in a distributive manner, showed a high level of
specificity for 3' terminal U's and was blocked by a terminal dU or pCp;
there was a slight exonucleolytic activity on a 3' terminal A or C, but no
activity on a 3' terminal G residue. The enzyme preferred single-stranded
oligo[U] 3' and did not digest duplex RNA. Two other 3'-5' exoribonuclease
activities were also detected in the mitochondrial extract, one of which
was stimulated by a 3'-phosphate and the other of which degraded RNAs with
a 3'OH to mononucleotides in a processive manner. The properties of the
distributive U-specific 3'-5' exoribonuclease suggest an involvement in
the U-deletion RNA editing reaction that occurs in the mitochondrion of
these cells.
This work was performed by Ruslan Aphasizhev.
Guide RNAs of the recently isolated LEM 125 strain of
Leishmania tarentolae: an unexpected complexity
Guide RNAs (gRNAs) are encoded both in the maxicircle and minicircle
components of the mitochondrial DNA of trypanosomatid protozoa. These RNAs
mediate the precise insertion and deletion of U residues in transcripts of
the maxicircle DNA. We showed previously that the old UC laboratory strain
of Leishmania tarentolae apparently lost more than 40 minicircle-encoded
gRNAs which are present in the recently isolated LEM125 strain (Thiemann
et al., 1994). We have further analyzed the population of
minicircle-encoded gRNAs in the LEM125 strain. Sau3AI and MspI minicircle
libraries were constructed and screened for novel gRNAs by negative colony
hybridization. This search yielded 20 minicircles encoding new gRNAs that
covered most of the remaining gaps in the editing cascades of the ND8,
ND9, G3 and G4 genes, and in addition, more than 30 minicircles containing
either unassigned or undetectable gRNA genes. We also completely sequenced
34 of the 45 minicircle sequence classes encoding previously identified
gRNAs. A total of 19 pairs of redundant gRNAs, which are gRNAs of
different sequences covering the same editing blocks, were identified. The
redundant gRNA pairs showed a differential steady-state abundance and
mismatches may represent one of the factors determining this abundance
differences. Alignments of the minicircles encoding redundant gRNAs
yielded 59 to 93% matching nucleotides, suggesting an origin from
duplication of ancestral minicircles and subsequent genetic drift. We
propose a functional explanation for the existence of redundant gRNAs in
this strain.
Differential localization of nuclear-encoded tRNAs
between the cytosol and mitochondrion in Leishmania tarentolae
All mitochondrial tRNAs of the kinetoplastid protozoan Leishmania
tarentolae are encoded in the nucleus and are imported from the cytosol
into the mitochondrion. We previously reported the partitioning of five
tRNAs and found that all were shared between the two compartments to
different extents. In order to increase our knowledge of the tRNAs of this
organism, and to attempt to understand the signals involved in their
subcellular localization, a method to RT-PCR amplify new tRNAs was
developed. Various tRNAs were 3' polyadenylated and reverse transcribed
with a sequence tagged primer. The cDNA was tagged by ligation to an
anchor oligonucleotide, and the resulting double-tagged cDNA was amplified
by PCR. Four new tRNAs were obtained, bringing to 20 the total number of
L. tarentolae tRNAs identified to date. The subcellular localization of 17
tRNAs was quantitatively analyzed by two-dimensional gel electrophoresis
and Northern hybridization. In general, the previously suggested
operational classification of tRNAs into three groups (mainly cytosolic,
mainly mitochondrial, and shared between the two compartments) is still
valid, but the relative abundance of each tRNA in the cytosol or
mitochondrion varies greatly as does the level of expression.
This work was performed by Steve Kapushoc and Juan Alfonzo.
Trypanosome Mitochondrial 3' Terminal Uridylyl
Transferase (TUTase): A Key Enzyme in U-insertion/deletion RNA Editing
A 3' terminal RNA uridylyltransferase was purified from mitochondria of
Leishmania tarentolae and the gene cloned and expressed from this species
and from Trypanosoma brucei. The enzyme is specific for 3' U-addition in
the presence of Mg++. TUTase is present in vivo in at least two stable
configurations: One contains a ~500 kDa TUTase oligomer and the other a
~700 kDa TUTase complex. Anti-TUTase antiserum specifically
co-precipitates a small portion of the p45 and p50 RNA ligases and
approximately 40% of the guide RNAs. Inhibition of TUTase expression in
procyclic T. brucei by RNAi down-regulates RNA editing and appears to
affect parasite viability.
This work was performed by Ruslan Aphasizhev et al.
Modification of the universally unmodified uridine-33 in
a mitochondria-imported edited tRNA and the role of the anticodon arm
structure on editing efficiency
Editing of tRNA has a wide phylogenetic distribution among eukaryotes
and in some cases serves to expand the decoding capacity of the target
tRNA. We previously described C to U editing of the wobble position of the
imported tRNATrp in Leishmania mitochondria which is essential for
decoding UGA codons as tryptophan. Here we show the complete set of
nucleotide modifications in the anticodon arm of the mitochondrial and
cytosolic tRNATrp as determined by electrospray ionization mass
spectrometry. This analysis revealed extensive mitochondria-specific
post-transcriptional modifications, including the first example of
thiolation of U33, the "universally unmodified" uridine. In light of the
known rigidity imparted on sugar conformation by thiolation, our discovery
of a thiolated U33 suggests that conformational flexibility is not a
universal feature of the anticodon structural signature. In addition, the
in vivo analysis of tRNATrp variants presented shows a single base pair
reversal in the anticodon stem of tRNATrp is sufficient to abrogate
editing in vivo, indicating that subtle changes in anticodon structure can
have drastic effects on editing efficiency.
This work was performed by Crain, P.F., Alfonzo, J.D., Rozenski, J.,
Kapushoc, S.T., McCloskey, J.A. and Simpson, L.
Wobble modification differences and subcellular
localization of tRNAs in Leishmania tarentolae: implication for tRNA
sorting mechanism
In Leishmania tarentolae, all the tRNAs required for mitochondrial
translation are encoded in the nuclear genome and imported from the
cytosol. It is known that tRNAGlu(UUC) and tRNAGln(UUG) are localized in
both cytosol and mitochondria. We investigated structural differences
between affinity-isolated cytosolic (cy) and mitochondrial (mt) tRNAs for
Glu and Gln by direct enzymatic sequencing and mass spectrometric
analysis. A unique modification difference in both tRNAs was identified at
the anticodon wobble position: cy tRNAs have
5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), whereas mt tRNAs have 5-
methoxycarbonylmethyl-2'-O-methyluridine (mcm5Um). In addition, a trace
portion (4%) of cy tRNAs was found to have 5-methoxycarbonylmethyluridine
(mcm5U) at its wobble position, which could represent a common
modification intermediate for both modified uridines in cy and mt tRNAs.
We also isolated a trace amount of mitochondria-specific tRNALys(UUU) from
the cytosol and found mcm5U at its wobble position, while its
mitochondrial counterpart has mcm5Um. We found that mt tRNALys and in
vitro-transcribed tRNAGlu were imported much more efficiently into
isolated mitochondria than the native cy tRNAGlu in an in vitro
importation assay, indicating that cytosol specific 2-thiolation could
play an inhibitory role in tRNA import into mitochondria.
This work is the result of a collaboration and was performed by Tomonori
Kaneko, Takeo Suzuki, Stephan T. Kapushoc, Mary Anne Rubio, Jafar
Ghazvini, Kimitsuna Watanabe and Tsutomu Suzuki
A 100 kDa complex of two RNA-binding proteins from
mitochondria of Leishmania tarentolae catalyzes RNA annealing and
interacts with several RNA editing components
A stable 100 kD complex from mitochondria of Leishmania tarentolae
containing two RNA-binding proteins, Ltp26 and Ltp28, was identified by
cross-linking to unpaired 4-thiouridine nucleotides in a partially duplex
RNA substrate. The genes were cloned and expressed and the complex was
reconstituted from recombinant proteins in the absence of RNA or
additional factors. The Ltp26 and Ltp28 proteins are homologues of gBP27
and gBP29 from Crithidia fasciculata, and gBP25 and gBP21 from Trypanosoma
brucei, respectively. The purified Ltp26/Ltp28 complex, the individual
recombinant proteins and the reconstituted complex are each capable of
catalyzing the annealing of complementary RNAs, as was previously shown
for gBP21 from T. brucei. A high molecular weight RNP complex consisting
of the Ltp26/Ltp28 complex and several 55-60 kDa proteins together with
guide RNA could be purified from mitochondrial extract of L. tarentolae
transfected with Ltp28-TAP. This complex also interacted in a less stable
manner with the RNA ligase-containing L- complex and with the 3' TUTase.
The Ltp26/Ltp28 RNP complex is a candidate for catalyzing the annealing of
guide RNA and pre-edited mRNA in the initial step of RNA editing.
Update 6-16-2003: These proteins are now labeled MRP1 and MRP2 and the
complex is labeled MRP1/2.
This work was performed by R. Aphasizhev, I. Aphasizhev and R. Nelson.
Is the Trypanosoma brucei REL1 RNA ligase specific for
U-deletion RNA editing and the REL2 RNA ligase specific for U-insertion
editing?
It was shown previously that the REL1 mitochondrial RNA ligase in
Trypanosoma brucei was a vital gene and disruption affected RNA editing in
vivo whereas the REL2 RNA ligase gene could be down regulated with no
effect on cell growth or on RNA editing. We performed down regulation of
REL1 in procyclic T. brucei (midgut insect forms) by RNAi and found a
40-50% inhibition of Cyb editing, which has only U-insertions, as well as
a similar inhibition of ND7 editing, which has both U-insertions and
U-deletions. In addition, both U-insertion and U-deletion in vitro
pre-cleaved editing were inhibited to similar extents. We also found
little if any effect of REL1 down regulation on the sedimentation
coefficient or abundance of the RNA ligase-containing L-complex
(Aphasizhev et al. (2003) EMBO J. 22:913-924), suggesting that the
inhibition of both insertion and deletion editing was not due to a
disruption of the L-complex. Together with the evidence that down
regulation of REL2 has no effect on cell growth or on RNA editing in vivo
or in vitro, these data suggest that the REL1 RNA ligase may be active in
vivo in both U-insertion and U-deletion editing. The in vivo biological
role of REL2 remains obscure.
This work was performed by G. Gao.
Isolation of an RNA Editing Complex Active in Both
U-insertion and U-deletion in vitro Editing
A multiprotein, high molecular weight complex active in both U-insertion
and U-deletion as judged by a pre-cleaved RNA editing assay was isolated
from mitochondrial extracts of Leishmania tarentolae by the tandem affnity
purification (TAP) procedure, using three different TAP-tagged proteins of
the complex. This editing- or E-complex consists of at least three
protein-containing components interacting via RNA: the RNA
ligase-containing L-complex, a 3' TUTase (terminal uridylyltransferase)
and two RNA-binding proteins, Ltp26 and Ltp28. Thirteen approximately
stoichiometric components were identified by mass spectrometric analysis
of the core L-complex: two RNA ligases; homologs of the four Trypanosoma
brucei editing proteins; and seven novel polypeptides, among which were
two with RNase III, one with an AP endo/exonuclease and one with
nucleotidyltransferase motifs. Three proteins have no similarities beyond
kinetoplastids.
This figure shows the isolated L-complex sediments as a single band in a
glycerol gradient and contains around 15 proteins.
This work was performed by Ruslan Aphasizhev, Inna Aphasizheva, Robert
E.Nelson, Guanghan Gao, Agda M.Simpson, Xuedong Kang, Arnold M.Falick, and
Sandro Sbicego
RBP38, an RNA-binding protein from trypanosomatid
mitochondria, modulates RNA stability
A novel RNA-binding protein, RBP38, was isolated from Leishmania
tarentolae mitochondria. This protein does not contain any known RNA
binding motifs and is highly conserved among the trypanosomatids but no
homologues were found in other organisms. Recombinant LtRBP38 binds single
and double stranded RNA substrates with dissociation constants in the 100
nanomolar range, as determined by fluorescence polarization analysis. Down
regulation of expression of the homologous gene, TbRBP38, in procyclic T.
brucei using conditional RNAi resulted in 80% reduction of steady-state
levels of RNAs transcribed from both maxicircle and minicircle DNA. In
organello pulse-chase labeling experiments were used to determine the
stability of RNAs in mitochondria that were depleted of TbRBP38. The
half-life of metabolically labeled RNA decreased from ~160 min to ~60 min
after depletion. In contrast, there was no change in transcriptional
activity. These observations suggest a role of RBP38 in stabilizing
mitochondrial RNA.
This work was performed by Sandro Sbicego, Juan D. Alfonzo, Antonio M.
Estévez, Mary Anne T. Rubio, Xuedong Kang, Christoph W. Turck and Marian
Peris
Genomic Organization of Trypanosoma brucei Kinetoplast
DNA Minicircles
The sequences of seven new Trypanosoma brucei kinetoplast DNA
minicircles were obtained. A detailed comparative analysis of these
sequences and those of the 18 complete kDNA minicircle sequences from T.
brucei available in the database was performed. These 25 different
minicircles contain 86 putative gRNA genes. The number of gRNA genes per
minicircle varies from 2 to 5. In most cases, the genes are located
between short imperfect inverted repeats, but in several minicircles there
are inverted repeat cassettes that did not contain identifiable gRNA
genes. Five minicircles contain single gRNA genes not surrounded by
identifiable repeats. Two pairs of closely related minicircles may have
recently evolved from common ancestors: KTMH1 and KTMH3 contained the same
gRNA genes in the same order, whereas KTCSGRA and KTCSGRB contained two
gRNA genes in the same order and one gRNA gene specific to each. All
minicircles could be classified into two classes on the basis of a short
substitution within the highly conserved region, but the minicircles in
these two classes did not appear to differ in terms of gRNA content or
gene organization. A number of redundant gRNAs containing identical
editing information but different sequences were present. The alignments
of the predicted gRNAs with the edited mRNA sequences varied from a
perfect alignment without gaps to alignments with multiple mismatches.
Multiple gRNAs overlapped with upstream gRNAs, but in no case was a
complete set of overlapping gRNAs covering an entire editing domain
obtained. We estimate that a minimum set of approximately 65 additional
gRNAs would be required for complete overlapping sets. This analysis
should provide a basis for detailed studies of the evolution and role in
RNA editing of kDNA minicircles in this species.
This work was done by Min Hong.
The RET1 TUTase adds U's to the gRNA 3' end and the RET2
TUTase adds U's at editing sites of mRNAs
We have described two TUTases, RET1 (RNA Editing TUTase 1) and RET2
(RNA Editing TUTase 2) as components of different editing complexes.
TAP-tagged Trypanosoma brucei RET2 was expressed and localized to
the cytosol in Leishmania tarentolae cells by removing the
mitochondrial signal sequence. Double affinity isolation yielded tagged
TbRET2 together with a few additional proteins. This material exhibits a
U-specific transferase activity in which a single uridine is added to the
3' end of a single-stranded RNA, thereby confirming that RET2 is a 3'
TUTase.
We also found that RNA interference of RET2 expression in T. brucei
inhibits in vitro U-insertion editing and has no effect on the length of
the 3'-oligo[U] tails of the gRNAs, whereas down regulation of RET1 has a
minor effect on in vitro U-insertion editing but produces a decrease in
the average length of the oligo[U] tails.
This suggests that RET2 is responsible for U-insertions at editing sites
and RET1 is involved in gRNA 3'-end maturation, which is essential for
creating functional gRNAs. From these results we have functionally
relabelled the previously described TUT-II complex containing RET1 as the
GP-complex (Guide RNA Processing).
This work was done by Ruslan Aphasizhev and Inna Aphasizheva.
Loss of editing due to extended culture of cells.
Larry Simpson, Stephen M. Douglass, James A. Lake,Matteo Pellegrini and Feng Li
We analyzed by NGS technology the mitochondrial genomes and transcriptomes of two strains,
the old lab UC strain and the recently isolated LEM125 strain. PacBio sequencing provided
complete minicircle sequences which avoided the assembly problem of short reads caused
by the conserved regions. Minicircles were identified by a characteristic size, the presence
of three short conserved sequences, a region of inherently bent DNA and the presence of
single gRNA genes at a fairly defined location. The LEM125 strain contained over 114 minicircles encoding different gRNAs and the UC strain only ~24 minicircles. Some LEM125
minicircles contained no identifiable gRNAs. Approximate copy numbers of the different
minicircle classes in the network were determined by the number of PacBio CCS reads
that assembled to each class. Mitochondrial RNA libraries from both strains were mapped
against the minicircle and maxicircle sequences. Small RNA reads mapped to the putative
gRNA genes but also to multiple regions outside the genes on both strands and large RNA
reads mapped in many cases over almost the entire minicircle on both strands. These data
suggest that minicircle transcription is complete and bidirectional, with 3’ processing yielding the mature gRNAs. Steady state RNAs in varying abundances are derived from all maxicircle genes, including portions of the repetitive divergent region. The relative extents of
editing in both strains correlated with the presence of a cascade of cognate gRNAs.
U-insertion/deletion RNA editing multiprotein complexes and
mitochondrial ribosomes in Leishmania tarentolae are located
in antipodal nodes adjacent to the kinetoplast DNA
Richard G. Wong, Katelynn Kazane, Dmitri A. Maslov, Kestrel Rogers, Ruslan Aphasizhev and Larry Simpson
We studied the intramitochondrial localization of several multiprotein complexes involved in U-insertion/deletion
RNA editing in trypanosome mitochondria. The editing complexes are located in one or two antipodal nodes
adjacent to the kinetoplast DNA (kDNA) disk, which are distinct from but associated with the minicircle catenation
nodes. In some cases the proteins are in a bilateral sheet configuration. We also found that mitoribosomes have a
nodal configuration. This type of organization is consistent with evidence for protein and RNA interactions of
multiple editing complexes to form an ~40S editosome and also an interaction of editosomes with mitochondrial
ribosomes





 tarentolae
tarentolae