
Click here to
see an animated pan-editing of L. tarentolae A6 mRNA (must use MIE ver 5;
advance by clicking)
Purpose:
- This site is intended to act as a source of
information on the U insertion/deletion type of RNA editing and to
increase the interactions between researchers in this field.
Definition
of U-insertion/deletion RNA editing:
- The insertion and deletion of uridine
(U) residues, usually within coding regions of mRNA transcripts of
cryptogenes in the mitochondrial genome of kinetoplastid protozoa.
Definitions of Terms used
in U-insertion/deletion RNA Editing.
Sequence Database:
The Nomenclature Problem and a Guide to a Solution
Aphasizheva,
I., Alfonzo, J., Carnes, J., Cestari, I., Cruz-Reyes, J., Goringer, U.,
Hajduk, D., Lukes, J., Madison-Antenucci, S., Maslov, D., McDermont, S.,
Ochenreiter, T., Read, L., Salavati, R., Schnaufer, A., Simpson, L.,
Stuart, K., Yurchenko, V., Zhou, Z., Zikova, A., Zhang, L., Zimmer, S. and
Aphasizhev, R. (2020). Lexis and Grammar of Mitochondrial RNA Processing
in Trypanosomes. Trends in Parasitol. In Press. (PDF)
Sequence Maps, Diagrams and Some Interesting Results (with
my comments):
- Taxonomy
of kinetoplastid protozoa
- Diagram
of the single mitochondrion and the kDNA network in L. tarentolae
stationary phase cell
- Electron micrograph
of a fragment of a kDNA network from L. tarentolae
- Another electron micrograph
of a fragment of a kDNA network from L. tarentolae
- Several micrographs
of kDNA networks from L. tarentolae
- Map
of maxicircle of L. tarentolae showing localization of
genes, cryptogenes and gRNA genes
- Comparative maps
of maxicircles of several species.
- The pan-edited NADH
dehydrogenase 7 gene in T. brucei
(Koslowsky et al., 1990) (This is the cover photo that got a lot of
people interested in RNA editing!)
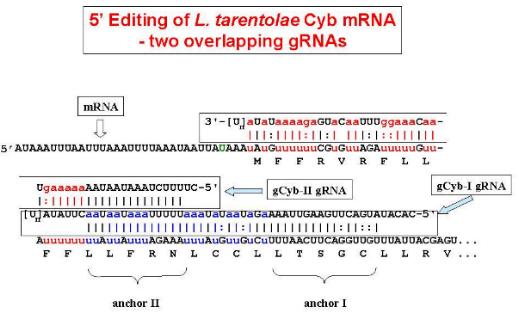
- Overlapping gRNAs
for the pan-edited A6 mRNA in L. tarentolae
- Overlapping gRNAs
for pan-edited RPS12 mRNA in L. tarentolae (Maslov
and Simpson, 1992)(Evidence for
independent editing of three domains)
- Comparative genomic organization
of gRNA genes in different kinetoplastids
- The
enzyme cascade and transesterification models for RNA editing
- Modified enzyme cascade model
for RNA editing (Byrne et al., 1996) (Development of a gRNA-medited in vitro U-insertion editing
system from L. tarentolae. Intermediates were visualized by RT-PCR.
The basic modification of the model is that U's are added in an
untemplated manner to the 3' end of the 5' cleavage fragment and then
deleted by an exonucleaase - in both deletion and insertion
situations)
- A
model for U-deletion (Seiwert
et al., 1996)(A breakthrough in
development of an in vitro gRNA-mediated U-deletion system in which A6
editing intermediates can be visualized on a gel. Provided evidence
against the transesterification model and for the enzyme cascade
model)
- A model for RNA editing in
kinetoplastids (Kable et al., 1996)(Application of the A6 in vitro editing system from T.
brucei for gRNA-mediated U-insertion editing)
- In vitro editing of site 1 of the A6 mRNA in T. brucei
- evidence for involvement of gRNA-mediated endonuclease, 3' to 5'
exonuclease and RNA ligase activities(An
elegant analysis of the in vitro system developed by Seiwert et al.)
- Sterochemical evidence supporting the enzyme cascade
model of U-insertion editing (Frech and Simpson, 1996)(Analyzed
the gRNA-independent U-insertion editing reaction in L. tarentolae.
The internal as well as the 3' incorporation of (SP)-alpha-S-UTP
proceeds via inversion of the stereoconfiguration. Consistent with the
enzyme cascade model and inconsistent with the transfer of U's from
the 3' end of the gRNA).
- Secondary structure
of a gRNA from T. brucei (Schmid et al.,
1995)
- Tertiary structure
of a gRNA from T. brucei (Hermann et al.,
1997)
- RNA binding domain
for the gRNA-binding protein from T. brucei, gBP21
(Hermann et al., 1997)
- Identification
of three riboendonuclease activities in mitochondrial extract from T.
brucei (Piller et al. 1997).(Identification of gRNA-independent and gRNA-dependent
cleavage activities)
- Disruption of a DEAD-Box gene in T. brucei
affects RNA
editing. (Missel et al., 1997)(First
demonstration that KO of a gene affects RNA editing)
- Purification of a functional enzymatic editing complex
from T. brucei mitochondria (Rusche et al., 1997) (This
20S complex consists of only 8 polypeptides, including three that may
represent RNA ligase components!)
- RNA editing
occurs in the free-living bodonid, Bodo saltans (Blom et al.,
1998)
- U-deletion editing differs enzymatically from
U-insertion editing (Cruz-Reyes et al., 1998) (interesting
observation that ATP and ADP stimulate U-deletion in vitro editing and
inhibit U-insertion in vitro editing! But these two activities
cosediment in the purified 20S editing complex (Cruz-Reyes
et al., 1998))
- A book on modification and editing of RNA, edited by
Grosjean and Benne.
- Trypanosoma brucei RBP16 Is a Mitochondrial
Y-box Family Protein with Guide RNA Binding Activity (Hayman and Read,
1999)
- The involvement of gRNA-binding protein gBP21 in RNA
editing- an in vitro and in vivo analysis (Lambert et al.,1999)
- A cis-acting A-U sequence element induces
kinetoplastid U-insertions (Brown et al., 1999)(Uses
a novel assay for in vitro U-insertion editing)
- Mapping contacts between gRNA and mRNA in trypanosome
RNA editing (Leung and Koslowsky, 1999) (evidence
that the 3' oligo[U] tail interacts with the preedited region of the
mRNA)
- Large minicircles in kDNA from Trypanosoma avium
(journal cover
photo) (Yurchenko et al., 1999)
- In vitro U-insertion using a L. tarentolae
mitochondrial extract mediated by cis-acting guide RNAs (Kapuchoc and
Simpson, 1999)
- Computer simulations of random segregation of kDNA
minicircles in trypanosomatids (Savill and Higgs, 1999) (The random segregation hypothesis was shown by computer
simulations to successfully explain the presence of a few major and
many minor minicircle sequence classes, the long survival time of a
few of these classes, and the fluctuations of sequence class copy
numbers over time!)
- A 'Constructive Neutral Evolution' theory for the
evolution of U-insertion editing (Stoltzfus, 1999)(see
also Simpson and Maslov, 1999, and Simpson, RNA Editing - An
Evolutionary Perspective, in The RNA World Second Edition, Cold Spring
harbor Laboratory Press, p 585-608, 1999, for other speculations on
the origin of RNA editing)
- Kinetoplastid RNA editing does not require the
terminal 3' hydroxyl of guide RNA but modifications to the gRNA
terminus can inhibit U-insertion (Burgess et al., 1999)
- mRNA editing and localization of gRNA genes in
mitochondria of Phytomonas serpens (Maslov et al., 1998)
- Deletions of the COIII and Cyb genes in the maxicircle
DNA of the respiratory deficient Phytomonas serpens (Maslov
et al., 1999) (Phytomonas could yet
become a model system for respiratory-deficient trypanosomes; they are
certainly easier to work with than bloodstream T. brucei!)
- The ATPase 6 cryptogene from human-infecting Leishmania
is very similar to that from the lizard-infecting L.
tarentolae (Brewster and Barker, 1999) (More
evidence that lizard Leishmania and mammalian Leishmania
are closely related and that the separate genus, Sauroleishmania,
should be discarded. See also Croan and Ellis, 1996; Croan et al.,
1997; Noyes et al., Parasitol. Today 14, 167, 1998)
- Knockout of the glutamate dehydrogenase gene from
bloodstream T. brucei has no effect on RNA editing (Estevez et
al., 1999)(Another good theory destroyed
(possibly) by a hard fact!)
- Characterization of a partially purified mitochondrial
RNA ligase from Leishmania tarentolae. The ligase can
preferentially join RNA molecules bridged by a complementary RNA, and
the ligation is negatively affected by a gap between the donor and
acceptor nucleotides. (Blanc et al., 1999).
- Phytomonas serpens maxicircle sequences -
unedited and edited - were added to the Edited
Sequences Database on Nov. 26, 1999.
- First direct evidence for the translation of unedited
and edited mRNAs in the mitochondrion of a trypanosomatid (Horvath et
al.a, 2000); (Horvath et al.b, 2000). (It is
nice to actually see evidence for something we all felt was the case.
A real breakthrough!)
- Mitochondrial minicircles in the free-living bodonid
Bodo saltans contain two gRNA gene cassettes and are not found in
large networks (Blom et al., 2000). (The first
example of a kinetoplastid species with noncatenated, gRNA
gene-containing minicircles, which implies that the creation of
minicircles and minicircle networks are separate evolutionary events).
- RNA-binding properties of the mitochondrial Y-box
protein RBP16 (Pelletier et al., 2000) (Another
in the growing list of gRNA-binding proteins, the genetic function of
which is still uncertain).
- Trypanosoma brucei guide RNA poly(U) tail formation is
stabilized by cognate mRNA (McManus et al., 2000). (A
model is proposed in which the purine-rich region of the cognate mRNA
protects the uridine tail from a uridine exonuclease activity that is
present within the complex. Two chromatographically distinct TUTase
activities were detected in mitochondrial extract.)
- A specific C to U nucleotide modification in the first
position of the anticodon of the nuclear-encoded and
mitochondrial-imported tryptophan tRNA in Leishmania tarentolae allows
the decoding the the mitochondrial UGA tryptophan codon (Alfonzo et
al., 1999) (The first evidence for C to U
editing in the mitochondrion of trypanosomatids).
I apologize for the long gap in these listings (an example
of benign neglect of a web site). I will try to update them as I
have time to do so.
Click here
to return to the main RNA Editing site.
This page created and maintained by Larry
Simpson



