Click on the gene to get the edited or unedited sequences and click on the name to get the 'map'. The sequences are in GCG format. To get the entire maxicircle sequence, click on 'maxicircle'.
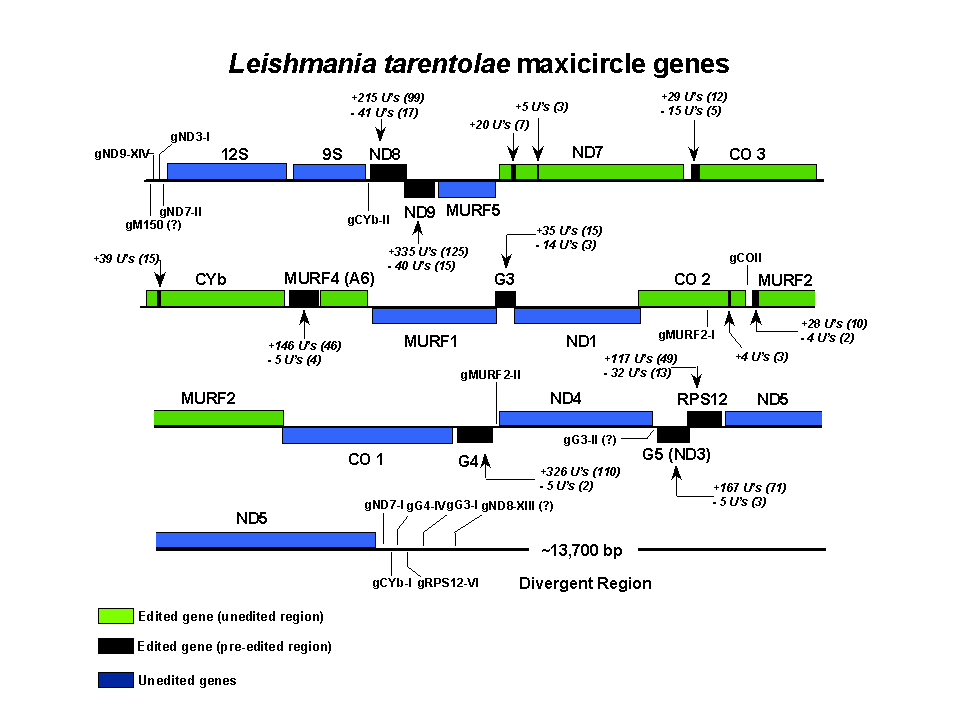
The maxicircle was linearized at the single EcoRI site. Genes above the line are 5' to 3', left to right, and genes below are 5' to 3', right to left. The number of U's added and deleted are indicated for each editing region, and the number of sites shown in parenthesis. The locations of maxicircle-encoded gRNAs are indicated and the gRNAs identified by gene and editing block (3' to 5' within the editing domain). The function of the divergent region, which consists of tandem AT-rich repeats of different complexities, is unknown. 12S and 9S are the mitochondrial rRNAs. CO = cytochrome oxidase, MURF = maxicircle unidentified reading frame, ND = NADH dehydrogenase, CYb = cytochrome b, A6 = ATPase subunit 6, RPS12 = ribosomal protein S12, G1-G5 = G-rich regions 1-5. The sequences obtained by clicking the genes are in GCG format, except for the maxicircle sequence which is in Genbank format. Click within the edited region to get the edited sequence and click within the unedited region to get the pre-edited sequence.
Click here to return to the RNA Editing site.
This page created and maintained by
L. Simpson
Last revised on
5/26/97.