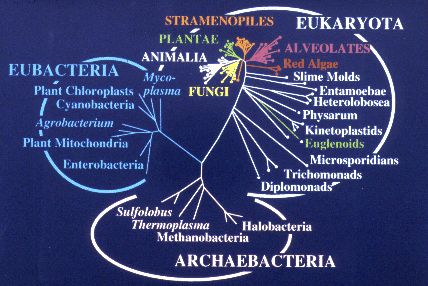
Ribosomal RNA sequences are among the slowest evolving gene sequences known and have proved very useful for molecular phylogeny studies of deeply diverged organisms. Phylogenetic reconstructions using SSrRNA sequences have showed that there are three major groups of organisms: eubacteria, archaebacteria and eukaryotes. Depending on the root of the Archaebacterial tree, there could be one other group which Lake calls the Eocytes (Sulfolobus). Note that the eukaryotic lineage consists of early diverged groups of unicellular protists and a "crown group" of metazoans - animals, plants and fungi.

Eukaryotes are distinguished among other things by having a double nuclear membrane, microtubules made of tubulin, microfilaments made of actin, the 9+2 flagellar and the 9 + 0 centriole structures, and a variety of membranellar organelles in the cytosol - the endoplasmic reticulum and the Golgi, and mitochondria and chloroplasts.
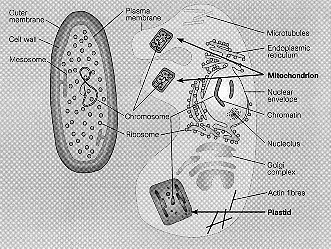
Mitochondria are double membrane organelles that contain the Kreb's cycle enzymes in the inner matrix, and the respiratory chain and the linked oxidative phosphorylation enzymes in theinner membrane. They also contain DNA and a complete transcriptrion and translation apparatus. The mitochondrial genome is much smaller than the nuclear genome and can contain as few as 3 genes.
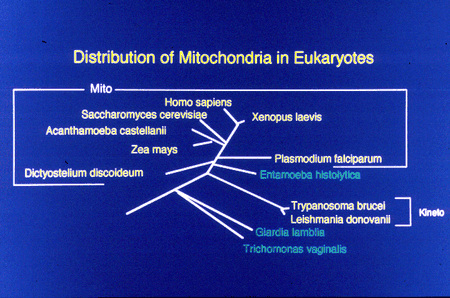
Mitochondria are found in all extant eukaryotes except a few lower protists - e.g. Giardia, Trichomonas, Entamoeba and a few ciliates.
This theory was first presented over a century ago, but received its best support and articulation by Lynn Margulis in 1970. The theory goes as follows:
This theory has almost achieved the staus of a paradigm. Most people agree that the molecular sequence data accumulated between 1975 and 1995 essentially confirmed the endosymbiosis theory for the origin of mitochondria. The remaining mitochondrial-encoded genes and nulcear-encoded genes for mitochondrial proteins were shown to be of alpha-proteobacterial origin, and the remaining nuclear genes were shown to be of archaebacterial origin.
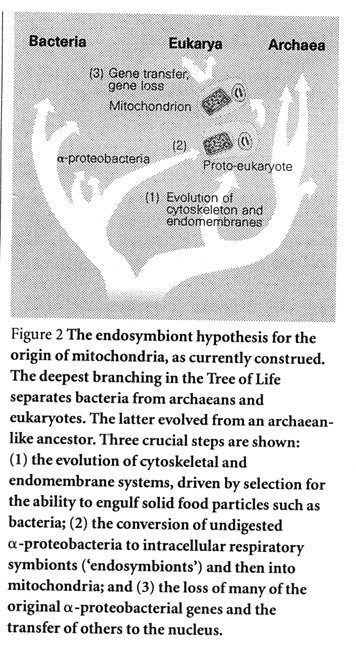 Taken
from
Doolittle (1998), Fig. 2. Taken
from
Doolittle (1998), Fig. 2. |
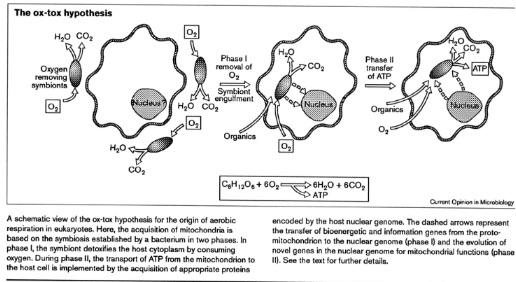 Taken from Andersson and Kurland, 1999. |
The mitochondrial genome codes for a limited number of RNAs and proteins essential for a functional mitochondrion. These include components of Complex I of the respiratory chain (NADH dehydrogenase), Complex II (succinate dehydrogenase), Complex III (cytrochrome b and c1), Complex IV (cytochrome oxidase), and Complex V (ATP synthetase). Two small rRNAs and, in animal and plant cells, a set of tRNAs, are also encoded in this DNA. In plants and protists, some of the proteins of the mitochondrial ribosomes are also encoded. Most of the genetic information for mitochondrial biogenesis is in the nuclear DNA, and the proteins are imported into the organelle by specific pathways. The size of the mitochondrial genome varies from 366 kb in Arabidopsis to 6 kb in Plasmodium. Human mitochondrial DNA encodes 13 proteins, 2 rRNAs and 22 tRNAs. Arabidopsis encodes only 2.5 fold more proteins as human.
There is a mitochondrial genome sequencing project ongoing which has already led to the complete sequences of the mt genomes from 23 protists which are at the root of the eukaryotic tree. The mitochondrial genome in R. americana, an early diverging protist, appears to be the closest to the genome of the presumed eubacterial endosymbiont. This DNA has a total of 97 genes, including all those found in other mitochondria and 18 genes novel to this organism. These 18 genes include foour genes for a eubacterial like multicomponent RNA polymerase. In all other mitochondria known the mt RNA polymerase is similar to that from T3 and T7 bacteriophage, and the origin of this is not known.
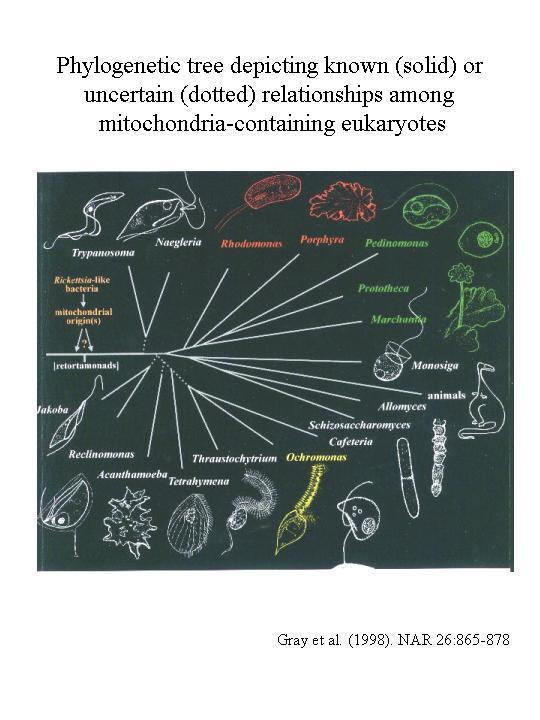
Did the endosymbiosis that produced the mitochodrion occur one time or multiple times? All the phylogenetic SSU rRNA and protein evidence support a single origin of mitochondria. For example, all of the genes found in all known mt DNAs are a subset of those found in R. americana mt DNA. This would be hard to reconcile with several independent reductions of larger eubacterial genomes. Furthermore all mt genes (both in the mt DNA and the nuclear DNA) tree with the alpha-proteobacteria.
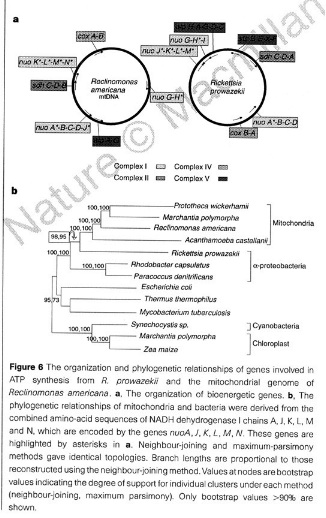
From
Andersson et al. (1998), Fig. 6.
The complete genome sequence of the obligate intracellular parasite, Rickettsia prowazekii, was recently published. This alpha-proteobacterium is the causal agent of louse borne typhus. The genome is 1,111,523 bp in size and contains 834 protein coding genes. A complete set of genes encoding TCA cycle and respiratory chain complex components was found. ATP production is the same as in modern mitochondria. Many genes involved in the biosynthesis and regulation of biosynthesis of amino acids and nucleosides present in free-living bacterial are absent in R. Prow and also in mitochondria. Loss of genes esential to a free-living existence makes the symbiosis obligatory.
This genome has the highest proportion of non-coding DNA (24%) so far detected in a microbial genome. These may be degraded remnants of neutralized genes that await elimination from the genome. This is an example of reductive evolution such as may have occurred in the evolution of the mitochondrion, but which probably occurred independently. Molecular phylogeny of several genes places this genome closest to modern mitochondrial genomes.
The Hydrogenosome
This organelle was first described by microscopists as a double membrane bound particle with little internal structure. In 1973 Muller described an isolation procedure using Trichomonas vaginalis that separated these particles from mitochondria. The name is derived from the function of producing hydrogen gas from the oxidation of pyruvate or malate. The organelle has a double membrane and the presence of DNA is still controversial but unlikely. These organelles are present in a variety of organisms living in anoxic oxygen environments such as free-living ciliates, rumen symbionts, and parasites, but few have been studied in any detail. Trichomonas does not have mitochondria and was initially thought to be one of the amitochondrial protists known as Archaezoa.
Glycolysis occurrs in the cytosol - glucose is oxidized to pyruvate with the aid of NAD+, with a net yield of 2 mol ATP. In mitochondrial eukaryotes pyruvate is further oxidized in themitochondrion through the pyruvate dehydrogenase complex (PDH), the Krebs Cycle and O2 respiration to yield CO2 and water together with theproduction of 36 mol ATP per mol glucose. In amitochondrial eukaryotes, pyruvate is metabolized through pyruvate:ferredoxin reductase (PFO) rather than PDH. Cytosolic pyruvate is importedinto the hydrogenosome and PFO coverts it into CO2, acetyl CoA and reduced ferredoxin. Ferredoxin is reoxidized by hydrogenase, yielding hydrogen. This metabolism yields 2 additional mol of ATP per mol glucose.
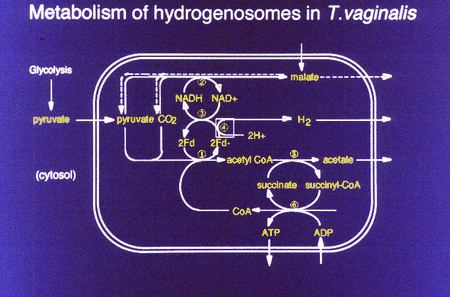

The evolutionary origin of the hydrogenosome was debated since its discovery. Recently several independent molecular studies have provided evidence that the hydrogenosome and the mitochondrion share evolutionary ancestry. (Germont et al., 1996; Bui et al., 1996; Roger et al, 1996).
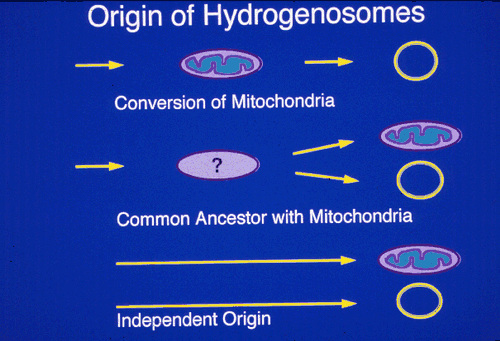
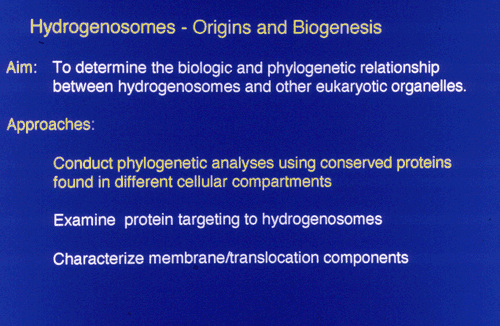
The molecular approaches used to determine the evolutionary origins of hydrogenosomes were as follows:
 |
 |
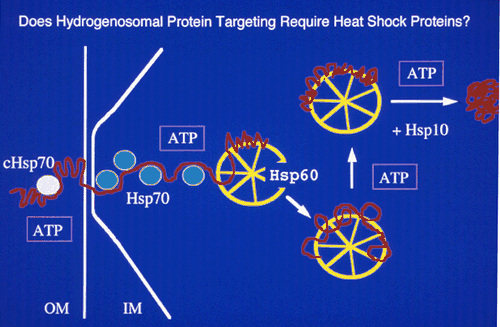
A typical protein import assay:
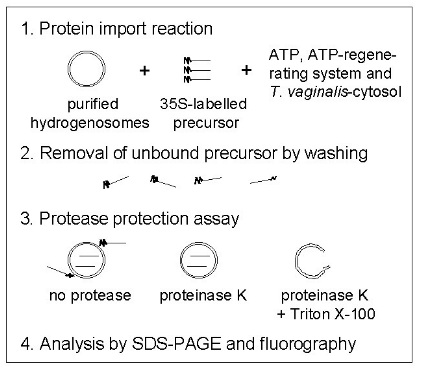
A second method used to identify the components of the protein import system:
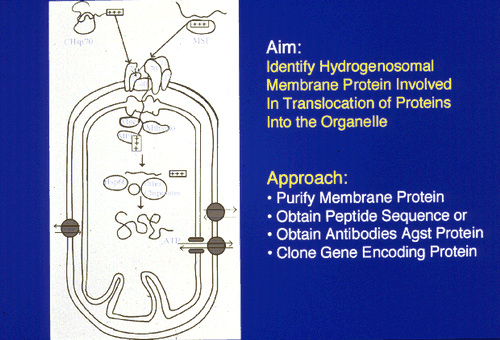
The Hsp60, Hsp70 and Hsp10 chaperonin proteins
which are known to be involved in the transport of
proteins into mitochondria were identified in
hydrogenosomal membranes. These proteins tree to
other mitochondrial and to proteo-bacterial
proteins.
Cleaved mitochondrial-like targeting signals were identified in these proteins. The mature forms lacked short N-terminal sequences.
This evidence suggested that hydrogenosomes and mitochondria share a common ancester.
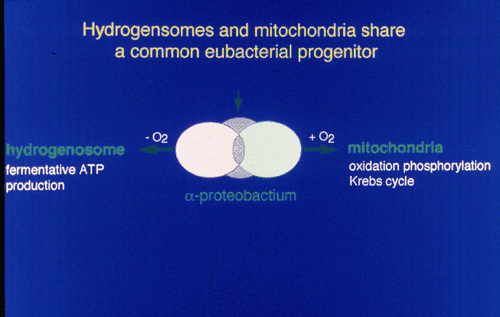 |
 |
This evidence indicates that modern amitochondrial eukaryotes lost their mitochondria during evolution.
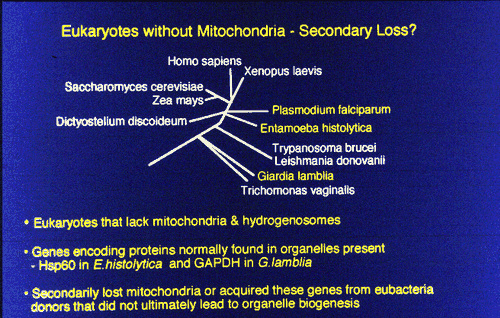
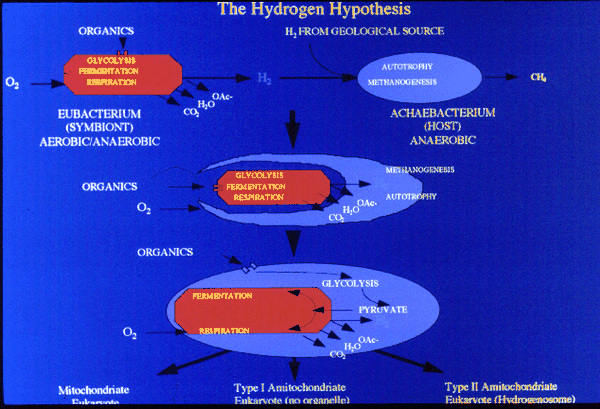
Martin and Muller (1998) proposed a novel theory for the origin of the mitochondrion - based on the work on the hydrogenosome and the demonstration that extant amitochondrial protists probably lost their mitochondria or have modified mitochondria. They claim that there was a nacient symbiosis between an alpha-proteobacterium and an Archael cell, but that the rlevant activity was not respiration but the excretionof H2 and CO2, which are the waste products of anerobic fermentation. The archaean host like the modern methane-producing achaeans, used H2 and CO2 as its sole source os of energy and carbon. The archaean host became dependent on the symbiont. Gene transfer from symbiont to host gave the host membrane proteins for substrate import and enzymes for glycolysis. The host was then able to feed the symbiont. The host was thereby converted from autotrophy (using H2 and CO2) to heterotrophy (using organic molecules from the environment). The symbiont was then either lost, converted to a hydrogenosome or became a mitochondrion.
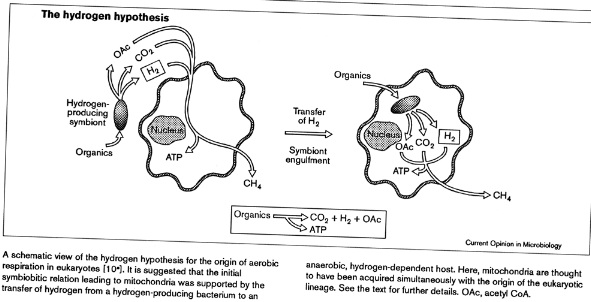
Taken from
Andersson and Kurland (1999)
A summary figure: