Gene Expression in African Trypanosomes
Nascent transcripts are obtained by the Run-On Assay
This assay was used to show that transcription was sensitive to α-amanatin
An indication that transcription of some genes in trypanosomes was by an unusual polymerase
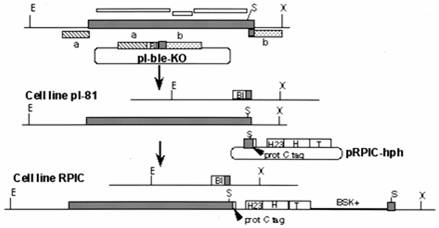
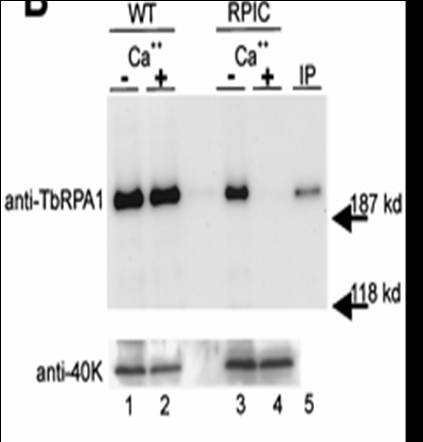
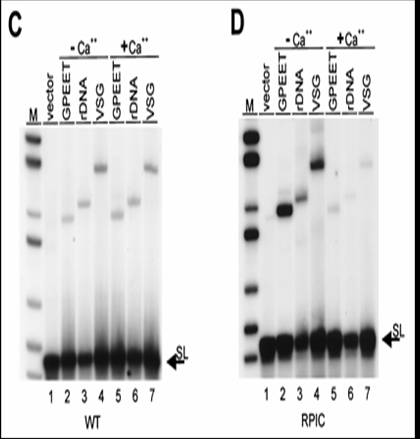
Gunzl et al (2003) showed by removal of the different polymerases by immunoprecipitation that VSGs were transcribed by Pol I and SL genes (see below) by Pol II. To obtain specific antibodies he first knocked out one allele of Pol I in T. brucei and then replaced the other allele with a protein C-tagged version. Protein C binds IgG at high affinity and the binding requires Ca++. The depletion of Pol I affected in vitro transcription of rRNA, the GPEET procyclin RNA and a VSG RNA. There was no affect on transcription of the SL gene, tubulin and heat shock protein 70 genes.
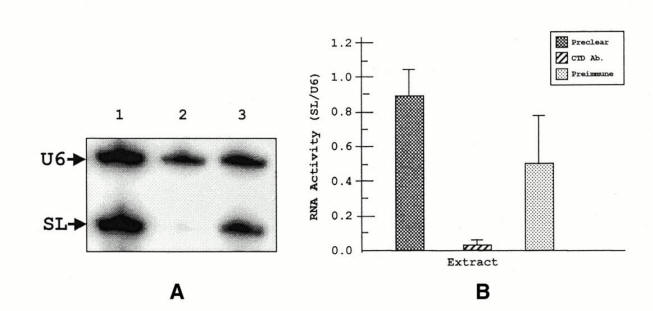
Early work using UV inactivation Kinetics was showed polycistronic transcription at the VSG locus
Early sequencing of the complete chromosome 1 of L. major showed that there were only two blocks of orfs oriented in opposite directions. This was consistent with previous evidence for polycistronic transcription units.
Almost complete genome sequences of the three trypanosomatids, T. brucei, L. major and T. cruzi, were published in 2005. n In T. brucei only the telomeres and minichromosomes were not completed. The above results for Leishmania chromosome 1 were amply confirmed in that both strands of the 11 megabase chromosomes in T. brucei contain long, non-overlapping gene clusters (directional gene clusters of DGCs). These are probably transcribed as polycistrons and the transcripts then trans-spliced and polyadenylated. In Leishmania major there are 133 clusters of ten to several hundred protein-coding genes on t he same strand. the genes are not related in functions. The clusters span u p to 1259 kb and are separated by 900 bp to 14 kb divergent "strand-switch" regions. Transcription initiates and terminates bidirectionally within these regions. The direction of transcrption is towards the telomeres.
The Tr-Tryps have all the shared and conserved subunits of RNAP I, II and III, except for two subunits. Very few conserved transcription regulators were found, which is consistent with the known fact that transcription is not regulated and that control is mainly post-transcriptional.
RNA pol subunits and transcription factors in trypanosomatids:
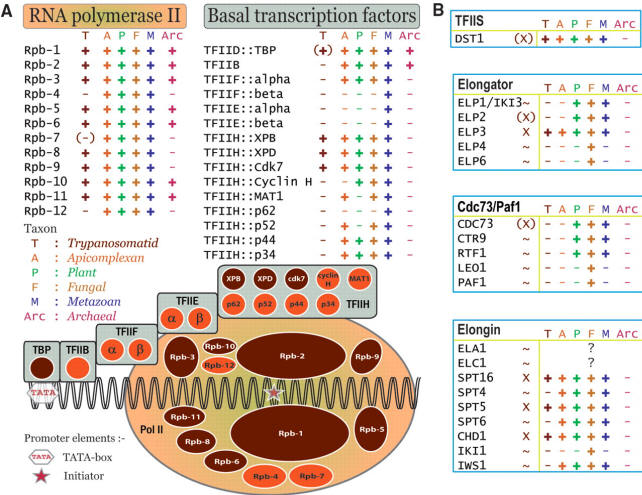
Figure from Ivens et al. (2005).
Trans-splicing
A recent review of the role of the three RNA polymerases in trypanosomes is available here.
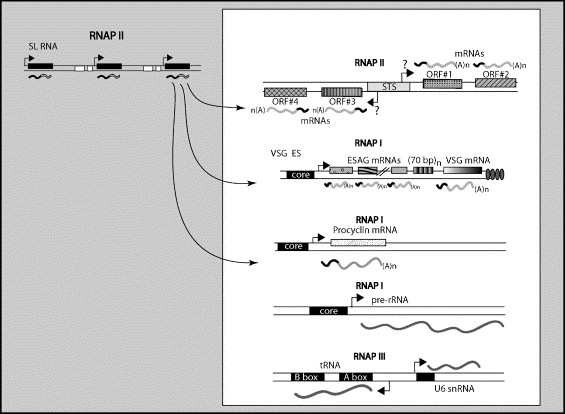
The purpose of trans-splicing of the spliced leader sequence is to provide a highly modified pre-made cap to the 5' end of the transcripts. 3' polyadenylation of the first gene in a tandem array occurs after the second gene gets the SL cap on its 5' end. The genomes of T. brucei, T. cruzi and L. major are known. Homologues are present for subunits of RNAP II but not for many of the known transcription factors such as TFIIB,TFIIF, TFIIE, some TFIIH subunits, the largest subunit of TFIIA and the TATA biding protein associated factors (TAFs). This could be due to divergence of the sequences but also possibly to the lack of transcriptional regulation in trypanosomes.
RNA polymerase II
Most information about RNAP II in trypanosomes has come from the study of the SL RNA gene transcription. So far this is the only defined RNAP-II dependent promoter known. There is no TATA box or B-recognition element (BRE). There is a proximal bi-partite sequence element (PSE) that is similar to the human U1 snRNA promoter. This element however only has 3 of the normal 5 subunits. Highly diverged homologues of TFIIA and TRB copurify with the SNAP. These factors are associated not only with the 5' end but also the 3' untranslated sequences, unlike all other systems.
RNA polymerase I
In most cells RNAP I transcribes pre-rRNA but in T. brucei it is also used to transcribe the VSG genes and the insect stage-specific EP and GREET proteins. This is unique to T. brucei perhaps due to selective pressure from having to produce a lot of the VSG surface proteins to escape the immune response.
RNA polymerase III
This polymerase transcribes tRNAs and U-rich snRNAs. A unique aspect of the trypanosome system is that some of the RNAP III promoters also act as promoter elements for U6, U3 and 7SL RNA genes on the other strand.
Post Transcriptional Regulation of Gene Expression
AU-rich sequence motifs in the 3' untranslated regions of mRNAs (AREs) appear to be involved in regulating the stability of the molecules. Specific RNA binding proteins were shown to bind to these AREs and stimulate a 3' to 5' exonuclease activity in the "exosome". Due to the apparent absence of transcriptional regulation, posttranscriptional regulation of mRNA stability by trans-splicing and decapping and 3' AREs plays a major role in trypanosomes.
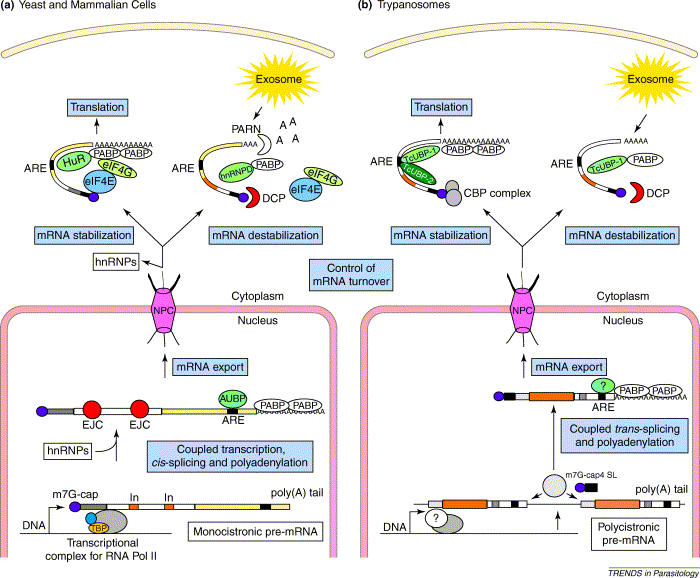
The Exosome
The exosome is a complex of 10 or so proteins that are involved in degradation of RNA. The exosome has been studied in T. brucei by Estevez et al. (2003). See figure below from this paper.
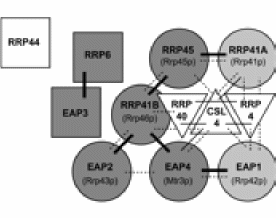
Summary schematic depicting the T. brucei exosome interactions from the two-hybrid analysis (panel A) and from the co-depletion experiments (Figs. 2 and 3). RNase PH-like subunits are represented as circles, S1 domain proteins are shown as triangles, and the remaining subunits are represented as squares. The names of the RNase PH-like human homologues are shown in parentheses. Two-hybrid interactions detected in both directions are shown in thick solid lines; those detected only in one direction are shown in thin solid lines. TbEAP3 and TbRRP40 self-interactions are not shown. The interactions described elsewhere for the components of the human and yeast exosomes are shown in dashed lines. Those T. brucei subunits for which depletion results in co-depletion of both TbRRP4 and TbRRP45 are filled in dark gray; the subunits for which depletion results in co-depletion of only TbRRP4 are filled in light gray, and those subunits for which depletion does not cause co-depletion of neither TbRRP4 or TbRRP45 are shown as empty shapes.
From D'Orso et al. (2003). Comparison of RNA-processing mechanisms in (a) yeast and mammalian cells and (b) trypanosomes. The overall steps (blue boxes) forming part of mRNA maturation processes (after transcription) are shown in both cell types. Molecules forming part of the different RNA processing mechanisms are indicated. The grey circle in the nucleus of trypanosome cells indicates the presence of a trans-splicing and polyadenylation machinery. Abbreviations: A, adenylic acid; ARE, AU-rich element; AUBP, AU-rich RNA-binding protein; CBP, cap-binding complex; DCP, decapping complex; eIF, eukaryotic translation initiation factor; EJC, exon-junction complex; hnRNP, heteronuclear ribonucleoprotein; HuR, human RNA-binding protein; In, intron; m7G-cap, tri-methyl-guanosine or CAP structure; mRNA, messenger RNA; NPC, nuclear pore complex; PABP, poly(A)-binding protein; PARN, poly(A)-ribonuclease; RNA Pol II, RNA polymerase II; SL, spliced leader; TcUBP, Trypanosoma cruzi U-rich-binding protein; TBP, TATA-binding protein.
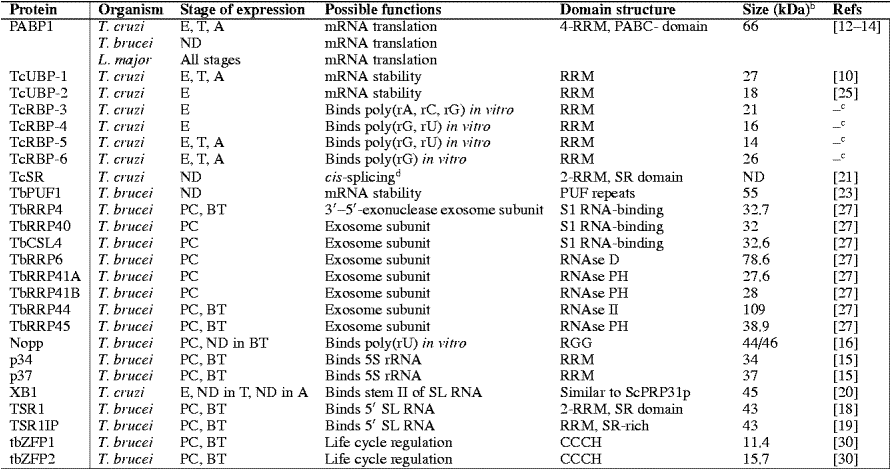
Major RNA binding proteins in trypanosomes:
Taken from same paper.
Questions:
1. What are two novel features of transcription in trypanosomes?
2. Why do you think th at it has been so difficult to identify a Pol II promoter in trypanosomes?
3. What type of genetic regulation is used in trypanosomes - transcriptional, post transcriptional or translational?
4. Why is the trans splicing of the SL a possible target for chemotherapy?
5. How is mRNA turnover regulated in trypanosomes?