Serum Sensitivity
The natural immunity of humans to the cattle pathogen, Trypanosoma brucei, but not to the morphologically indistinguishable human pathogen, T. rhodesiense, is due to the selective non-immune lysis of the former parasite by a factor or factors in normal human serum (NHS). Trypanosome Lytic Factor (TLF) consists of two classes of particles; TLF1 is a subclass of high density lipoprotein (HDL) and TLF-2 is a high molecular weight complex. These factors can be separated by gel filtration. There is some controversy as to the precise nature of the lytic factors in human serum.
TLF-1 are 18 nm high density particles that contain phospholipids, cholesterol, cholesterol esters, and apolipoproteins. Drain et al. (2001) found that TLF1 has a high affinity binding site on T. brucei and that binding is mediated through a haptoglobin (Hp)-like receptor. The evidence was that Hpr shows competition for TLF1 binding and a monoclonal antibody against Hpr prevents TLF1 uptake and trypanosome lysis.

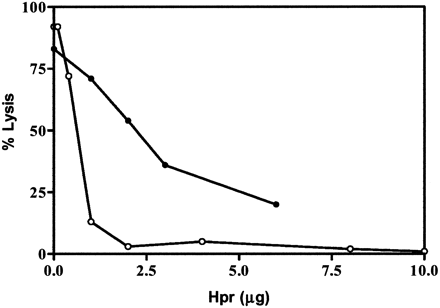
Dose-dependent inhibition of TLF-1-mediated lysis by Hpr and Hp. TLF-1 (4 units) was added to a standard TLF lysis assay with increasing amounts of purified Hpr (closed circles) or Hp (open circles). Cells were incubated for 2 h at 37 °C, and the percentage of lysed trypanosomes was determined by microscopic examination.
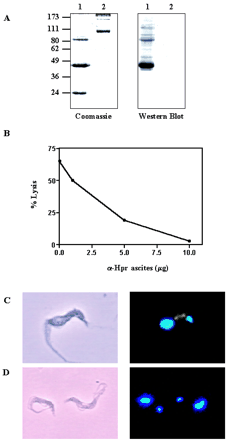
Inhibition of TLF-1 binding and uptake by an Hpr-specific
antibody. A, TLF-1 (lane 1) or purified human Hp (lane 2)
were separated by nonreduced 12% SDS-PAGE and Coomassie-stained or transferred
onto nitrocellulose. The Western blot was immunostained with a monoclonal
antibody specific for Hpr and detected by alkaline phosphatase. B, TLF-1
(1.5 units) was added to a standard TLF-1 assay and increasing amounts of Hpr
antibody. C, trypanosomes were incubated in the presence of 50 µg/ml
Alexa-488-conjugated TLF-1 for 1 h at 37 °C (green). Phase contrast and
fluorescence microscopy were performed on cells following methanol fixation. The
kinetoplast and nucleus were stained with Hoecht dye (blue). D,
trypanosomes were incubated in the presence of 50 µg/ml Alexa-488-conjugated
TLF-1 and 40 µg of monoclonal Hpr antibody and treated as in C.
These data led to the model that a haptoglobin-like component of
HDL is the TLF1 factor. The Hajduk lab also showed that TLF1 is taken up by endocytosis and targeted to a
lysosome-like compartment. The proposed
uptake pathway is indicated in the figure below.
Evidence indicates that the trypanolytic activity is due to peroxidase
activity of a ‘haptoglobin’-hemoglobin complex that causes oxidative damage to
the vesicular/lysosomal membrane releasing proteases etc. that subsequently
degrade the contents of the cytoplasm. Resistant T. brucei bind
the TLF1, but do not internalize it.
There is also a second Trypanosome Lytic Factor in Human Serum
Tomlinson et al. (1995) found that HDL-deficient serum from patients with Tangiers disease are as trypanolytic as serum from normal people, and that the lytic activity fractionated only in a high MW lipoprotein-depleted fraction. Two peaks of lytic activity were observed in both NHS and Tangier serum. They concluded that there must be second lytic factor not associated with HDL and called this TLF2.
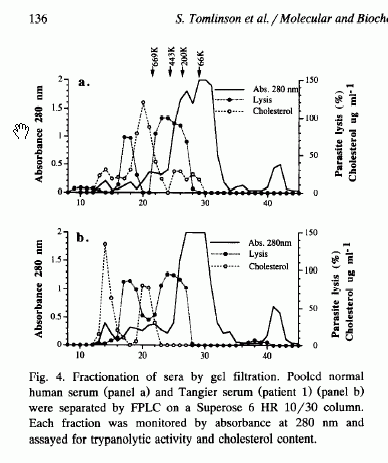
Raper et al. (1996) showed that the trypanolytic activity of purified TLF1
was inhibited by exogenous haptoglobin (Hp) at concentrations (0.1 mg/ml) lower
than those present in NHS (0.2-2 mg/ml), but Hp (up to 2.5 mg/ml) had no effect
on the lytic activity of either NHS or isolated TLF2. Serum depleted of Hp
showed higher lytic activity than NHS. They concluded that endogenous Hp
inhibits TLF1 activity, and that TLF2 is the main trypanolytic factor in NHS.
The story gets more complex
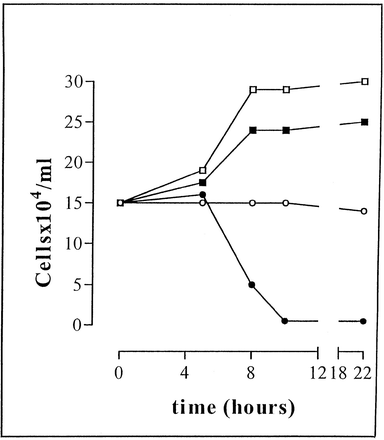
In a series of papers, the Raper and other labs lab showed that human TLF1 is composed of apoA-I, Hpr, apoL-I, cathelicid9 in antimirobial peptide (hCAP18), phospholipase D, apoA-II and paraoxonase. Molina-Portela et al (2005) have presented a new model for trypanolytic activty of serum that incorporates much of the above data. First they showed that human lytic serum promotes the influx of sodium ions into the parasite. This is followed by an influx of Cl- and influx of water. Pore formation in planar lipid bilayers by TLF1 was followed in real time.
They concluded that TLF1 generates cation-selective pores and that the apoL-I in TLF1 contributes by increasing the anion permeability. The components of TLF1 that could create pores are Hpr and hCAP18. This pore-forming mechanism is similar to a common immune defense strategy used by many organism to protect against pathogens.
The Study of Resistance to Human Serum - Another Approach to the Problem
The human serum resistance gene or SRA was discovered in a screen of cDNAs for mRNas expressed in resistant but not in sensitive lines of T. brucei. The SRA gene has been found in all T. rhodesiense isolates tested. In fact identification of this gene by PCR has proven to be a sensitive diagnostic method for human infective trypanosomes.
Van Xong et al. (1998) noted that resistance to lysis by NHS was associated with antigenic variation, and transcription of the SRA gene was found only in cells expressing the ETat 1.10 VSG. In fact the SRA gene turned out to be a expression gene associated gene or ESAG.

Direct evidence for the involvement of SRA in resistance to lysis was obtained by transfecting the SRA gene into sensitive T. brucei and showing that this conferred resistance to lysis.
Structure of the SRA Protein
Campillo and Carrington (2003) have modeled the SRA protein and found a relationship to the conserved VSG N-terminal domain fold structure, as shown below.
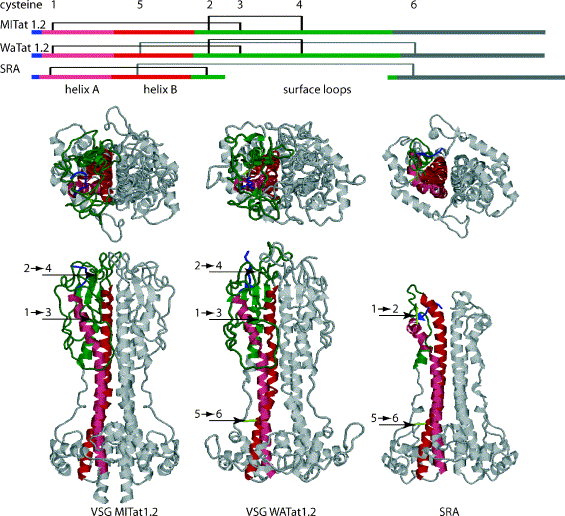
Comparison of the primary and tertiary structure of the N-terminal domains of
VSGs and SRA. Top: the primary structure of VSGs and SRA showing the location of
the N-terminus (blue), the two long
 -helices
A and B that form a coiled coil (pink and red) and the surface loops (green).
Bottom: the structure of the N-terminal domains of the MITat1.2 dimer and models
for the WATat1.2 and SRA dimers. The same color scheme in the primary and
tertiary structures and in each dimer only one monomer is color coded. In the
two models there are several small regions that could not be assigned a definite
structure, these are represented as differences between the two monomers. The
numbers indicate the locations of the cysteines in the primary sequence and the
location of the disulphide bridges in the tertiary structures and the cysteines
are indicated by the same numbers in
Fig. 1b. The PDB files for the models of WATat1.2 and SRA are available as
supplementary files.To obtain accurate models a sequence alignment using a
structure-assisted approach was produced [33].
The models were constructed using the following steps: (i) A structure-based
sequence alignment of the template structures (MITat1.2 and ILTat1.24) was
obtained using the COMPARER suite of programs [34
and
35] and formatted using the program JOY [36].
(ii) From the alignment, a sequence profile was constructed using the sequence
alignment program CLUSTALW [37].
(iii) The VSGs and subsequently the SRA sequence were then aligned to this
profile. Manual adjustment of the multiple alignment sequence was achieved using
the program Seaview [38].
(iv) From the alignment of SRA with the template VSGs, structural models
containing all non-hydrogen atoms were obtained automatically using the method
implemented in MODELLER [39].
The model corresponding to the lowest value of the objective function calculated
by MODELLER was used for further analysis. The model was validated using the
VERIFY 3D [40],
PROCHECK [41]
and COMPARER programmes. The models had to satisfy most restraints used to
calculate them, particularly those concerning their stereochemistry. (v) Cycles
of primary sequence realignment, modeling and structure validation were repeated
until no further improvement on the structure was observed. The final alignment
used to generate the structures is available as a supplementary file. Evaluation
of the final models with VERIFY 3D produced all positive scores. The average
score obtained with VERIFY 3D is 0.65 comparable with 0.70 for the
experimentally determined structure of MITat1.2. Evaluation of the
stereo-chemical quality of the model with PROCHECK showed that only a few
residues (2.2%) are in disallowed regions in Ramachandran plot.
-helices
A and B that form a coiled coil (pink and red) and the surface loops (green).
Bottom: the structure of the N-terminal domains of the MITat1.2 dimer and models
for the WATat1.2 and SRA dimers. The same color scheme in the primary and
tertiary structures and in each dimer only one monomer is color coded. In the
two models there are several small regions that could not be assigned a definite
structure, these are represented as differences between the two monomers. The
numbers indicate the locations of the cysteines in the primary sequence and the
location of the disulphide bridges in the tertiary structures and the cysteines
are indicated by the same numbers in
Fig. 1b. The PDB files for the models of WATat1.2 and SRA are available as
supplementary files.To obtain accurate models a sequence alignment using a
structure-assisted approach was produced [33].
The models were constructed using the following steps: (i) A structure-based
sequence alignment of the template structures (MITat1.2 and ILTat1.24) was
obtained using the COMPARER suite of programs [34
and
35] and formatted using the program JOY [36].
(ii) From the alignment, a sequence profile was constructed using the sequence
alignment program CLUSTALW [37].
(iii) The VSGs and subsequently the SRA sequence were then aligned to this
profile. Manual adjustment of the multiple alignment sequence was achieved using
the program Seaview [38].
(iv) From the alignment of SRA with the template VSGs, structural models
containing all non-hydrogen atoms were obtained automatically using the method
implemented in MODELLER [39].
The model corresponding to the lowest value of the objective function calculated
by MODELLER was used for further analysis. The model was validated using the
VERIFY 3D [40],
PROCHECK [41]
and COMPARER programmes. The models had to satisfy most restraints used to
calculate them, particularly those concerning their stereochemistry. (v) Cycles
of primary sequence realignment, modeling and structure validation were repeated
until no further improvement on the structure was observed. The final alignment
used to generate the structures is available as a supplementary file. Evaluation
of the final models with VERIFY 3D produced all positive scores. The average
score obtained with VERIFY 3D is 0.65 comparable with 0.70 for the
experimentally determined structure of MITat1.2. Evaluation of the
stereo-chemical quality of the model with PROCHECK showed that only a few
residues (2.2%) are in disallowed regions in Ramachandran plot.
Finally, to close the circle,
Vanhamme et al. (2003) showed that the N-terminal
a-helix of SRA is responsible for resistance to NHS and that this domain
interacted strongly with a C-terminal α-helix of the human apoL-I protein. They
also showed that SRA was localized to the lysosomal compartment.
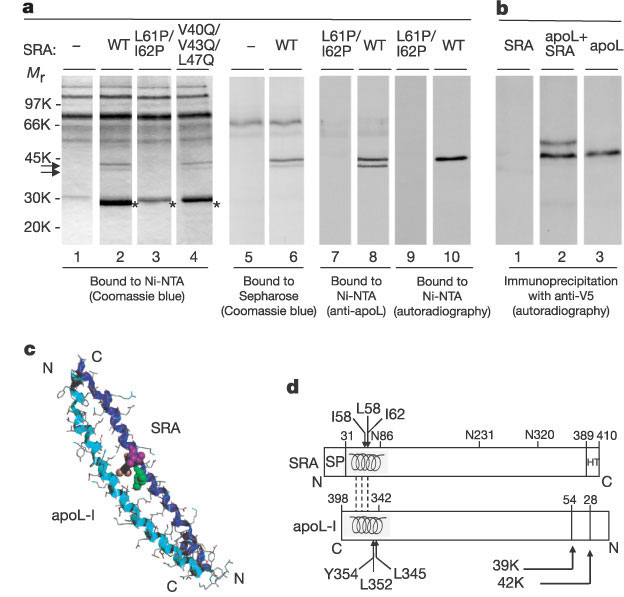
Binding of apoL-I to SRA. a, Patterns of NHS proteins bound to resins
containing either no protein (- ), or different versions of His-tagged SRA. NHS
(lanes 1–8) or in vitro synthesized [35S]-labelled apoL-I
(lanes 9, 10) was incubated with the resin. Bound proteins were eluted with
either imidazole (lanes 1–4, 7–10) or deoxycholate (lanes 5, 6), and revealed by
Coomassie blue staining (lanes 1–6), binding of anti-apoL-I antibodies (lanes 7,
8) or autoradiography (lanes 9, 10). The double arrow designates bands
specifically bound with functional SRA, and asterisks label eluted His-tagged
SRA. b, Immunoprecipitation with anti-V5 antibodies, of in vitro
synthesized [35S]-labelled SRA (48K) incubated with or without [35S]-labelled
V5–apoL-I (42K). The predicted [35S]-labelled methionine ratio
between these proteins is 4:9. c, Model of the SRA 31–79 fragment (dark
blue ribbon) in anti-parallel interaction with the apoL-I 340–392 fragment
(light blue ribbon). I58 (green), L61 (pink) and I62 (mauve) are in CPK
representation. d, SRA and apoL-I features mentioned in the text (SP,
signal peptide; HT, hydrophobic tail; broken lines symbolize the interactions
between the ![]() -helices
in each molecule).
-helices
in each molecule).
Depletion of apoL-I by incubation with SRA or antiserum led to a total loss of lytic activity. Addition of recombinant apoL-I restored lytic activity. They also showed that apoL-I is taken up by the parasite into the lysosomal compartment. The conclusion was that SRA in the lysosomal compartment binds to apoL-I and inhibits its lytic activity.
Questions:
1. What determines whether a trypanosome can infect humans?
2. What are the factors in human serum that lyse sensitive trypanosomes?
3. Discuss the controversy over the precise nature of the factors in NHS that lyse trypanosomes.
4. How did the study of the SRA protein lead to the identification of a specific component of NHS that is responsible for lysis?